8CX4
 
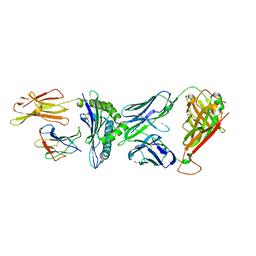 | | TCR-antigen complex AS8.4-YEIH-HLA*B27 | | Descriptor: | AS8.4a, AS8.4b, Beta-2-microglobulin, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
8CTG
 
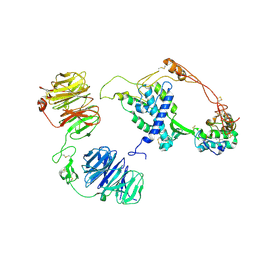 | | Extracellular architecture of an engineered canonical Wnt signaling ternary complex | | Descriptor: | Frizzled-8, Low-density lipoprotein receptor-related protein 6, PALMITOLEIC ACID, ... | | Authors: | Tsutsumi, N, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-14 | | Release date: | 2023-03-15 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Wnt-Frizzled-LRP6 initiation complex reveals the basis for coreceptor discrimination.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DH9
 
 | |
8DHA
 
 | |
8DH8
 
 | |
8DA3
 
 | | Coevolved affibody-Z domain pair LL1.c1 | | Descriptor: | Affibody LL1.FILF, Immunoglobulin G-binding protein A, MALONATE ION, ... | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DA9
 
 | | Coevolved affibody-Z domain pair LL2.c3 | | Descriptor: | Affibody LL2.FIIV, GLYCEROL, Immunoglobulin G-binding protein A, ... | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DA5
 
 | | Coevolved affibody-Z domain pair LL1.c4 | | Descriptor: | GLYCEROL, Immunoglobulin G-binding protein A, affibody LL1.FIVM | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DA8
 
 | | Coevolved affibody-Z domain pair LL2.c1 | | Descriptor: | Affibody LL2.FIIK, GLYCEROL, Immunoglobulin G-binding protein A | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DAC
 
 | | Coevolved affibody-Z domain pair LL2.c22 | | Descriptor: | Affibody LL2.FILV, GLYCEROL, Immunoglobulin G-binding protein A | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DA4
 
 | | Coevolved affibody-Z domain pair LL1.c2 | | Descriptor: | Affibody LL1.FIVM, Immunoglobulin G-binding protein A, SULFATE ION, ... | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DA6
 
 | | Coevolved affibody-Z domain pair LL1.c5 | | Descriptor: | Affibody LL1.FIIM, Immunoglobulin G-binding protein A | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DA7
 
 | | Coevolved affibody-Z domain pair LL1.c6 | | Descriptor: | Immunoglobulin G-binding protein A, MALONATE ION, affibody LL1.FIFV | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DAA
 
 | | Coevolved affibody-Z domain pair LL2.c7 | | Descriptor: | Affibody LL2.FIVK, Immunoglobulin G-binding protein A, MALONATE ION | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DAB
 
 | | Coevolved affibody-Z domain pair LL2.c17 | | Descriptor: | Affibody LL2.IVVY, Immunoglobulin G-binding protein A | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.134 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DZ8
 
 | |
8ENT
 
 | | Interleukin-21 signaling complex with IL-21R and IL-2Rg | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Abhiraman, G.C, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-09-30 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | A structural blueprint for interleukin-21 signal modulation.
Cell Rep, 42, 2023
|
|
8EWY
 
 | | Structure of Janus Kinase (JAK) dimer complexed with cytokine receptor intracellular domain | | Descriptor: | ADENOSINE, ADENOSINE-5'-DIPHOSPHATE, Interferon lambda receptor 1, ... | | Authors: | Caveney, N.A, Saxton, R.A, Waghray, D, Garcia, K.C. | | Deposit date: | 2022-10-24 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural basis of Janus kinase trans-activation.
Cell Rep, 42, 2023
|
|
4MS8
 
 | | 42F3 TCR pCPB9/H-2Ld Complex | | Descriptor: | 42F3 alpha, 42F3 beta, H-2 class I histocompatibility antigen, ... | | Authors: | Birnbaum, M.E, Adams, J.J, Garcia, K.C. | | Deposit date: | 2013-09-18 | | Release date: | 2014-09-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Structural interplay between germline interactions and adaptive recognition determines the bandwidth of TCR-peptide-MHC cross-reactivity.
Nat. Immunol., 17, 2016
|
|
4MQS
 
 | | Structure of active human M2 muscarinic acetylcholine receptor bound to the agonist iperoxo | | Descriptor: | 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Muscarinic acetylcholine receptor M2, Nanobody 9-8 | | Authors: | Kruse, A.C, Ring, A.M, Manglik, A, Hu, J, Hu, K, Eitel, K, Huebner, H, Pardon, E, Valant, C, Sexton, P.M, Christopoulos, A, Felder, C.C, Gmeiner, P, Steyaert, J, Weis, W.I, Garcia, K.C, Wess, J, Kobilka, B.K. | | Deposit date: | 2013-09-16 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nature, 504, 2013
|
|
4LDO
 
 | | Structure of beta2 adrenoceptor bound to adrenaline and an engineered nanobody | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, Camelid Antibody Fragment, L-EPINEPHRINE, ... | | Authors: | Ring, A.M, Manglik, A, Kruse, A.C, Enos, M.D, Weis, W.I, Garcia, K.C, Kobilka, B.K. | | Deposit date: | 2013-06-24 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Adrenaline-activated structure of beta 2-adrenoceptor stabilized by an engineered nanobody.
Nature, 502, 2013
|
|
4LDE
 
 | | Structure of beta2 adrenoceptor bound to BI167107 and an engineered nanobody | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Camelid Antibody Fragment, ... | | Authors: | Ring, A.M, Manglik, A, Kruse, A.C, Enos, M.D, Weis, W.I, Garcia, K.C, Kobilka, B.K. | | Deposit date: | 2013-06-24 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Adrenaline-activated structure of beta 2-adrenoceptor stabilized by an engineered nanobody.
Nature, 502, 2013
|
|
4LDL
 
 | | Structure of beta2 adrenoceptor bound to hydroxybenzylisoproterenol and an engineered nanobody | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 4-[(1R)-1-hydroxy-2-{[1-(4-hydroxyphenyl)-2-methylpropan-2-yl]amino}ethyl]benzene-1,2-diol, Camelid Antibody Fragment, ... | | Authors: | Ring, A.M, Manglik, A, Kruse, A.C, Enos, M.D, Weis, W.I, Garcia, K.C, Kobilka, B.K. | | Deposit date: | 2013-06-24 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Adrenaline-activated structure of beta 2-adrenoceptor stabilized by an engineered nanobody.
Nature, 502, 2013
|
|
4N0C
 
 | | 42F3 TCR pCPE3/H-2Ld complex | | Descriptor: | 42F3 VmCh alpha, 42F3 VmCh beta, H-2 class I histocompatibility antigen, ... | | Authors: | Birnbaum, M.E, Adams, J.J, Garcia, K.C. | | Deposit date: | 2013-10-01 | | Release date: | 2015-08-19 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural interplay between germline interactions and adaptive recognition determines the bandwidth of TCR-peptide-MHC cross-reactivity.
Nat. Immunol., 17, 2016
|
|
4N5E
 
 | | 42F3 TCR pCPA12/H-2Ld complex | | Descriptor: | 42F3 alpha VmCh, 42F3 beta VmCh, H-2 class I histocompatibility antigen, ... | | Authors: | Birnbaum, M.E, Adams, J.J, Garcia, K.C. | | Deposit date: | 2013-10-09 | | Release date: | 2015-08-19 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (3.059 Å) | | Cite: | Structural interplay between germline interactions and adaptive recognition determines the bandwidth of TCR-peptide-MHC cross-reactivity.
Nat. Immunol., 17, 2016
|
|
