7VX5
 
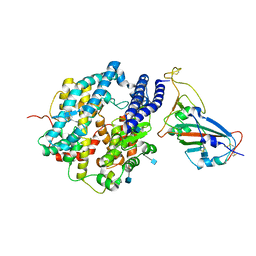 | | ACE2-RBD in SARS-CoV-2 Kappa variant S-ACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXD
 
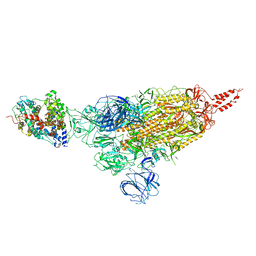 | | SARS-CoV-2 spike protein in complex with ACE2, Beta variant, C1 state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VX4
 
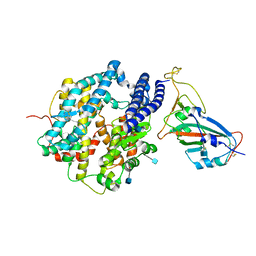 | | ACE2-RBD in SARS-CoV-2 Beta variant S-ACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXF
 
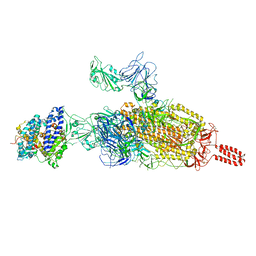 | | SARS-CoV-2 spike protein in complex with ACE2, Beta variant, C2B state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-01 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXE
 
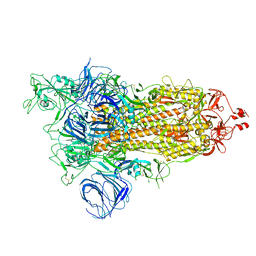 | | SARS-CoV-2 Kappa variant spike protein in open state | | Descriptor: | Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7WK2
 
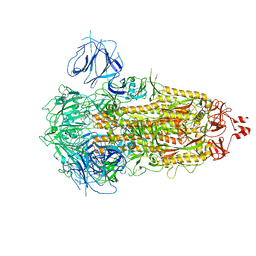 | | SARS-CoV-2 Omicron S-close | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-01-08 | | Release date: | 2022-01-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
7WKA
 
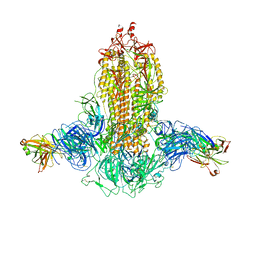 | |
7WK9
 
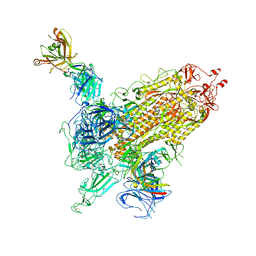 | |
7WK8
 
 | |
7WVO
 
 | | SARS-CoV-2 Omicron S-open-2 | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2022-03-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
7WVN
 
 | | SARS-CoV-2 Omicron S-open | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2022-03-02 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
7VXC
 
 | | SARS-CoV-2 Kappa variant spike protein in C3 state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXI
 
 | | SARS-CoV-2 Kappa variant spike protein in transition state | | Descriptor: | Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-22 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXK
 
 | | SARS-CoV-2 spike protein in complex with ACE2, Beta variant, C2A state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXB
 
 | | SARS-CoV-2 Kappa variant spike protein in C2b state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-22 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXM
 
 | | SARS-CoV-2 spike protein in complex with ACE2, Beta variant, C3 state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2022-01-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7WEV
 
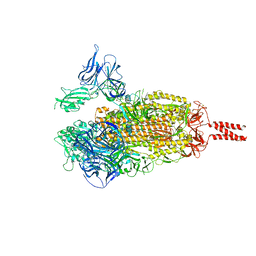 | | SARS-COV-2 BETA VARIANT SPIKE PROTEIN IN TRANSITION STATE | | Descriptor: | Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-12-24 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7WK3
 
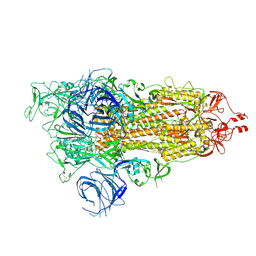 | | SARS-CoV-2 Omicron S-open | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-01-08 | | Release date: | 2022-01-26 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for ACE2 engagement and antibody evasion and neutralization of SARS-Co-2 Omicron varient
To Be Published
|
|
7V5G
 
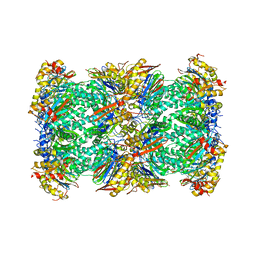 | | 20S+monoUb-CyclinB1-NT (S1) | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-08-17 | | Release date: | 2021-09-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | The 20S as a stand-alone proteasome in cells can degrade the ubiquitin tag.
Nat Commun, 12, 2021
|
|
7V5M
 
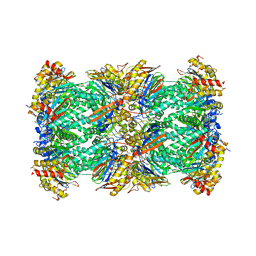 | | 20S+monoUb-CyclinB1-NT (S2) | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-08-17 | | Release date: | 2021-09-22 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | The 20S as a stand-alone proteasome in cells can degrade the ubiquitin tag.
Nat Commun, 12, 2021
|
|
4U7P
 
 | | Crystal structure of DNMT3A-DNMT3L complex | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Wang, L, Guo, X, Li, J, Xiao, J, Yin, X, He, S, Wang, J, Xu, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.821 Å) | | Cite: | Structural insight into autoinhibition and histone H3-induced activation of DNMT3A
Nature, 517, 2015
|
|
4U7T
 
 | | Crystal structure of DNMT3A-DNMT3L in complex with histone H3 | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Guo, X, Wang, L, Yin, X, Li, J, Xiao, J, He, S, Wang, J, Xu, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insight into autoinhibition and histone H3-induced activation of DNMT3A
Nature, 517, 2015
|
|
7LQ7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CV503 and COVA1-16 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-02-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern.
Sci Transl Med, 13, 2021
|
|
8ZCA
 
 | | Crystal structure of human CD47 ECD bound to Fab of Hu1C8 | | Descriptor: | 1C8 Fab heavy chain, 1C8 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Z, Ding, J. | | Deposit date: | 2024-04-29 | | Release date: | 2025-05-07 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An anti-CD47 antibody binds to a distinct epitope in a novel metal ion-dependent manner to minimize cross-linking of red blood cells.
J.Biol.Chem., 2025
|
|
8D36
 
 | |
