7ZHR
 
 | |
7ZHH
 
 | | Complex structure of drosophila Unr CSD789 and a poly(A) RNA sequence | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), SULFATE ION, Upstream of N-ras, ... | | Authors: | Hollmann, N.M, Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-04-06 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Upstream of N-Ras C-terminal cold shock domains mediate poly(A) specificity in a novel RNA recognition mode and bind poly(A) binding protein.
Nucleic Acids Res., 51, 2023
|
|
5A7X
 
 | | negative stain EM of BG505 SOSIP.664 in complex with sCD4, 17b, and 8ANC195 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB OF BROADLY NEUTRALIZING ANTIBODY 17B, FAB OF BROADLY NEUTRALIZING ANTIBODY 8ANC195, ... | | Authors: | Scharf, L, Wang, H, Gao, H, Chen, S, McDowall, A, Bjorkman, P. | | Deposit date: | 2015-07-10 | | Release date: | 2015-08-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Broadly Neutralizing Antibody 8Anc195 Recognizes Closed and Open States of HIV-1 Env.
Cell(Cambridge,Mass.), 162, 2015
|
|
5CFH
 
 | |
6FIB
 
 | | Structure of human 4-1BB ligand | | Descriptor: | Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor ligand superfamily member 9,4-1BBL -CH/CL fusion, Tumor necrosis factor ligand superfamily member 9,Uncharacterized protein | | Authors: | Joseph, C, Claus, C, Ferrara, C, von Hirschheydt, T, Prince, C, Funk, D, Klein, C, Benz, J. | | Deposit date: | 2018-01-17 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tumor-targeted 4-1BB agonists for combination with T cell bispecific antibodies as off-the-shelf therapy.
Sci Transl Med, 11, 2019
|
|
8OLU
 
 | |
5M5E
 
 | | Crystal structure of a interleukin-2 variant in complex with interleukin-2 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Klein, C, Freimoser-Grundschober, A, Waldhauer, I, Stihle, M, Birk, M, Benz, J. | | Deposit date: | 2016-10-21 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cergutuzumab amunaleukin (CEA-IL2v), a CEA-targeted IL-2 variant-based immunocytokine for combination cancer immunotherapy: Overcoming limitations of aldesleukin and conventional IL-2-based immunocytokines.
Oncoimmunology, 6, 2017
|
|
7P5R
 
 | | Racemic protein crystal structure of lacticin Q from Lactococcus lactis | | Descriptor: | D-Lacticin Q, FORMIC ACID, Lacticin Q | | Authors: | Lander, A.J, Li, X, Baumann, P, Rizkallah, P, Luk, L.Y.P, Jin, Y. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Roles of inter- and intramolecular tryptophan interactions in membrane-active proteins revealed by racemic protein crystallography.
Commun Chem, 6, 2023
|
|
5CSG
 
 | |
5B0V
 
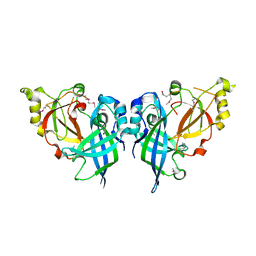 | | Crystal Structure of Marburg virus VP40 Dimer | | Descriptor: | ETHANOL, Matrix protein VP40 | | Authors: | Oda, S, Bornholdt, Z.A, Abelson, D.M, Saphire, E.O. | | Deposit date: | 2015-11-05 | | Release date: | 2016-01-20 | | Last modified: | 2016-03-09 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal Structure of Marburg Virus VP40 Reveals a Broad, Basic Patch for Matrix Assembly and a Requirement of the N-Terminal Domain for Immunosuppression
J.Virol., 90, 2015
|
|
5AMN
 
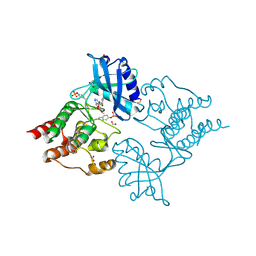 | | The Discovery of 2-Substituted Phenol Quinazolines as Potent and Selective RET Kinase Inhibitors | | Descriptor: | 4-[3-HYDROXYANILINO]-6,7-DIMETHOXYQUINAZOLINE, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Newton, R, Bowler, K, Burns, E.M, Chapman, P, Fairweather, E, Fritzl, S, Goldberg, K, Hamilton, N.M, Holt, S.V, Hopkins, G.V, Jones, S.D, Jordan, A.M, Lyons, A, McDonald, N.Q, Maguire, L.A, Mould, D.P, Purkiss, A.G, Small, H.F, Stowell, A, Thomson, G.J, Waddell, I.D, Waszkowycz, B, Watson, A.J, Ogilvie, D.J. | | Deposit date: | 2015-03-11 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The discovery of 2-substituted phenol quinazolines as potent RET kinase inhibitors with improved KDR selectivity.
Eur J Med Chem, 112, 2016
|
|
5CSB
 
 | | The crystal structure of beta2-microglobulin D76N mutant at room temperature | | Descriptor: | Beta-2-microglobulin | | Authors: | de Rosa, M, Mota, C.S, de Sanctis, D, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Conformational dynamics in crystals reveal the molecular bases for D76N beta-2 microglobulin aggregation propensity.
Nat Commun, 9, 2018
|
|
2KTX
 
 | | COMPLETE KALIOTOXIN FROM ANDROCTONUS MAURETANICUS MAURETANICUS, NMR, 18 STRUCTURES | | Descriptor: | KALIOTOXIN | | Authors: | Gairi, M, Romi, R, Fernandez, I, Rochat, H, Martin-Eauclaire, M.-F, Van Rietschtoten, J, Pons, M, Giralt, E. | | Deposit date: | 1997-02-27 | | Release date: | 1997-06-16 | | Last modified: | 2017-12-20 | | Method: | SOLUTION NMR | | Cite: | 3D structure of kaliotoxin: is residue 34 a key for channel selectivity?
J.Pept.Sci., 3, 1997
|
|
1PYP
 
 | |
5A8H
 
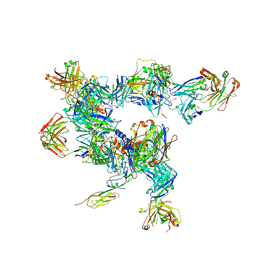 | | cryo-ET subtomogram averaging of BG505 SOSIP.664 in complex with sCD4, 17b, and 8ANC195 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB OF BROADLY NEUTRALIZING ANTIBODY 17B, FAB OF BROADLY NEUTRALIZING ANTIBODY 8ANC195 VARIANT G52K5, ... | | Authors: | Scharf, L, Wang, H, Gao, H, Chen, S, McDowall, A, Bjorkman, P. | | Deposit date: | 2015-07-15 | | Release date: | 2015-08-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Broadly Neutralizing Antibody 8ANC195 Recognizes Closed and Open States of HIV-1 Env.
Cell, 162, 2015
|
|
2K9I
 
 | |
5CS7
 
 | | The crystal structure of wt beta2-microglobulin at room temperature | | Descriptor: | Beta-2-microglobulin | | Authors: | de Rosa, M, Mota, C.S, de Sanctis, D, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational dynamics in crystals reveal the molecular bases for D76N beta-2 microglobulin aggregation propensity.
Nat Commun, 9, 2018
|
|
1KTX
 
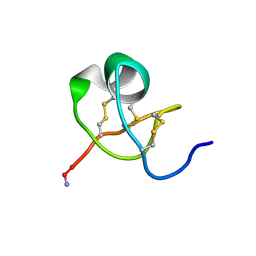 | | KALIOTOXIN (1-37) SHOWS STRUCTURAL DIFFERENCES WITH RELATED POTASSIUM CHANNEL BLOCKERS | | Descriptor: | KALIOTOXIN | | Authors: | Fernandez, I, Romi, R, Szendefi, S, Martin-Eauclaire, M.-F, Rochat, H, Van Rietschtoten, J, Pons, M, Giralt, E. | | Deposit date: | 1994-06-02 | | Release date: | 1995-01-26 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Kaliotoxin (1-37) shows structural differences with related potassium channel blockers.
Biochemistry, 33, 1994
|
|
2L1L
 
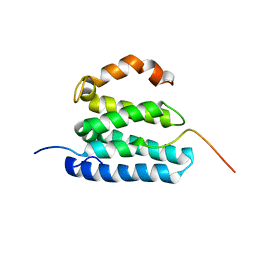 | |
4GFO
 
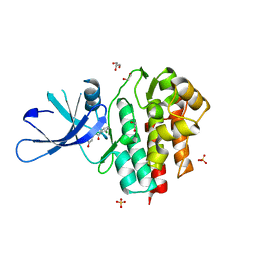 | | TYK2 kinase (JH1 domain) with 2,6-DICHLORO-N-(2-OXO-2,5-DIHYDROPYRIDIN-4-YL)BENZAMIDE | | Descriptor: | 1,2-ETHANEDIOL, 2,6-dichloro-N-(2-oxo-2,5-dihydropyridin-4-yl)benzamide, GLYCEROL, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2012-08-03 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lead identification of novel and selective TYK2 inhibitors.
Eur.J.Med.Chem., 67, 2013
|
|
4GJ3
 
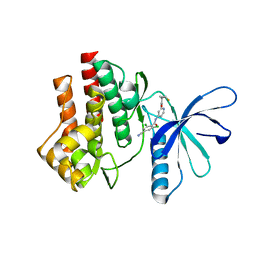 | | Tyk2 (JH1) in complex with 2,6-dichloro-4-cyano-N-[2-({[(1R,2R)-2-fluorocyclopropyl]carbonyl}amino)pyridin-4-yl]benzamide | | Descriptor: | 2,6-dichloro-4-cyano-N-[2-({[(1R,2R)-2-fluorocyclopropyl]carbonyl}amino)pyridin-4-yl]benzamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Ultsch, M.H. | | Deposit date: | 2012-08-09 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lead Optimization of a 4-Aminopyridine Benzamide Scaffold To Identify Potent, Selective, and Orally Bioavailable TYK2 Inhibitors.
J.Med.Chem., 56, 2013
|
|
6RLC
 
 | | Crystal structure of the PDZ tandem of syntenin in complex with fragment F13 | | Descriptor: | (2~{S})-2-[3-(4-chlorophenyl)sulfanylpropanoylamino]-3-methyl-butanoic acid, ACETATE ION, Syntenin-1 | | Authors: | Feracci, M, Barral, K. | | Deposit date: | 2019-05-02 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pharmacological inhibition of syntenin PDZ2 domain impairs breast cancer cell activities and exosome loadifing with syndecan and EpCAM cargo.
J Extracell Vesicles, 10, 2020
|
|
7ZDI
 
 | | PucB-LH2 complex from Rps. palustris | | Descriptor: | 1,2-Dihydro-psi,psi-caroten-1-ol, BACTERIOCHLOROPHYLL A, Light-harvesting protein, ... | | Authors: | Qian, P, Cogdell, R.J, Nguyen-Phan, T.C. | | Deposit date: | 2022-03-29 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of light-harvesting 2 complexes from Rhodopseudomonas palustris reveal the molecular origin of absorption tuning.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZE3
 
 | | PucD-LH2 complex from Rps. palustris | | Descriptor: | (3'E)-3',4'-didehydro-1,2-dihydro-psi,psi-caroten-1-ol, BACTERIOCHLOROPHYLL A, Light-harvesting protein B-800-850 alpha chain, ... | | Authors: | Qian, P, Cogdell, R.J, Nguyen-Phan, T.C. | | Deposit date: | 2022-03-30 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of light-harvesting 2 complexes from Rhodopseudomonas palustris reveal the molecular origin of absorption tuning.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8C3V
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.2-13 Fab and C1 nanobody | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, BA.2-13 heavy chain, BA.2-13 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-12-28 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
