1EVW
 
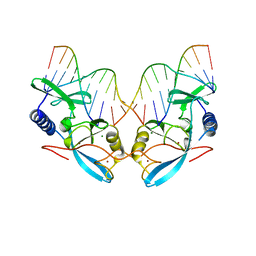 | | L116A MUTANT OF THE HOMING ENDONUCLEASE I-PPOI COMPLEXED TO HOMING SITE DNA. | | Descriptor: | DNA (5'-D(*TP*GP*AP*CP*TP*CP*TP*CP*TP*TP*AP*A)-3'), DNA (5'-D(*TP*GP*GP*CP*TP*AP*CP*CP*TP*TP*AP*A)-3'), DNA (5'-D(P*GP*AP*GP*AP*GP*TP*CP*A)-3'), ... | | Authors: | Galburt, E.A, Jurica, M.S, Chevalier, B.S, Erho, D, Stoddard, B.L. | | Deposit date: | 2000-04-20 | | Release date: | 2000-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformational changes and cleavage by the homing endonuclease I-PpoI: a critical role for a leucine residue in the active site.
J.Mol.Biol., 300, 2000
|
|
3E61
 
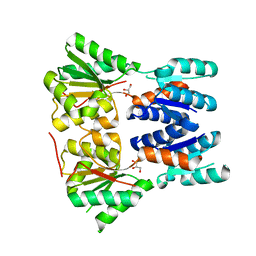 | | Crystal structure of a putative transcriptional repressor of ribose operon from Staphylococcus saprophyticus subsp. saprophyticus | | Descriptor: | GLYCEROL, Putative transcriptional repressor of ribose operon | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-14 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative transcriptional repressor of ribose operon from Staphylococcus saprophyticus subsp. saprophyticus
To be Published
|
|
6S20
 
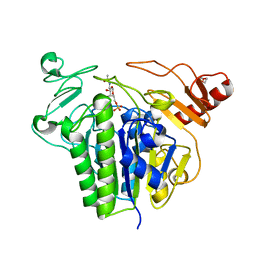 | | Metabolism of multiple glycosaminoglycans by bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus (BT33336S-sulf) | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-galactopyranose, CALCIUM ION, N-acetylgalactosamine-6-O-sulfatase, ... | | Authors: | Ndeh, D, Basle, A, Strahl, H, Henrissat, B, Terrapon, N, Cartmell, A. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Metabolism of multiple glycosaminoglycans by Bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus.
Nat Commun, 11, 2020
|
|
2MLD
 
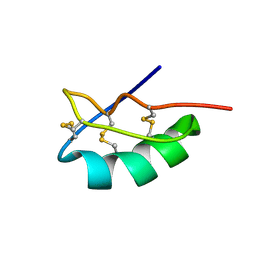 | |
3HAO
 
 | |
3E7J
 
 | | HeparinaseII H202A/Y257A double mutant complexed with a heparan sulfate tetrasaccharide substrate | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Heparinase II protein, ... | | Authors: | Shaya, D, Cygler, M. | | Deposit date: | 2008-08-18 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalytic mechanism of heparinase II investigated by site-directed mutagenesis and the crystal structure with its substrate.
J.Biol.Chem., 285, 2010
|
|
3VZ1
 
 | |
3GN4
 
 | | Myosin lever arm | | Descriptor: | CALCIUM ION, Calmodulin, MAGNESIUM ION, ... | | Authors: | Mukherjea, M, Llinas, P, Kim, H, Travaglia, M, Safer, D, Zong, A.B, Menetrey, J, Franzini-Armstrong, C, Selvin, P.R, Houdusse, A, Sweeney, H.L. | | Deposit date: | 2009-03-16 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Myosin VI dimerization triggers an unfolding of a three-helix bundle in order to extend its reach
Mol.Cell, 35, 2009
|
|
3ECA
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI L-ASPARAGINASE, AN ENZYME USED IN CANCER THERAPY (ELSPAR) | | Descriptor: | ASPARTIC ACID, L-asparaginase 2 | | Authors: | Swain, A.L, Jaskolski, M, Housset, D, Rao, J.K.M, Wlodawer, A. | | Deposit date: | 1993-07-02 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Escherichia coli L-asparaginase, an enzyme used in cancer therapy.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
2LVU
 
 | | Solution structure of Miz-1 zinc finger 10 | | Descriptor: | ZINC ION, Zinc finger and BTB domain-containing protein 17 | | Authors: | Bedard, M, Maltais, L, Beaulieu, M, Bernard, D, Lavigne, P. | | Deposit date: | 2012-07-11 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure note: solution structure of human Miz-1 zinc fingers 8 to 10.
J.Biomol.Nmr, 54, 2012
|
|
3VZ2
 
 | |
3V82
 
 | | Thaumatin by LB based Hanging Drop Vapour Diffusion after 1.81 MGy X-Ray dose at ESRF ID29 beamline (Best Case) | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Belmonte, L, Scudieri, D, Tripathi, S, Pechkova, E, Nicolini, C. | | Deposit date: | 2011-12-22 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Langmuir-Blodgett nanotemplate and radiation resistance in protein crystals: state of the art.
CRIT.REV.EUKARYOT.GENE EXPR., 22, 2012
|
|
6RK5
 
 | | Inter-dimeric interface controls function and stability of S-methionine adenosyltransferase from U. urealiticum | | Descriptor: | Methionine adenosyltransferase | | Authors: | Shahar, A, Zarivach, R, Bershtein, S, Kleiner, D, Shmulevich, F. | | Deposit date: | 2019-04-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase.
J.Mol.Biol., 431, 2019
|
|
3V8C
 
 | | Crystal structure of monoclonal human anti-rhesus D Fc IgG1 t125(yb2/0) double mutant (H310 and H435 in K) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, GLYCEROL, ... | | Authors: | Menez, R, Stura, E.A, Bourel, D, Siberil, S, Jorieux, S, De Romeuf, C, Ducancel, F, Fridman, W.H, Teillaud, J.L. | | Deposit date: | 2011-12-22 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Effect of zinc on human IgG1 and its Fc gamma R interactions.
Immunol.Lett., 143, 2012
|
|
6RJS
 
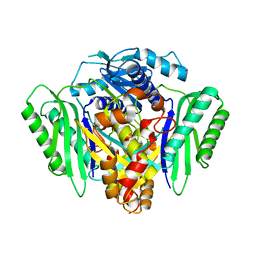 | | Inter-dimeric interface controls function and stability of S-methionine adenosyltransferase from U. urealiticum | | Descriptor: | Methionine adenosyltransferase | | Authors: | Shahar, A, Zarivach, R, Bershtein, S, Kleiner, D, Shmulevich, F. | | Deposit date: | 2019-04-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase.
J.Mol.Biol., 431, 2019
|
|
1EA7
 
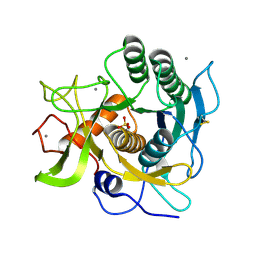 | | Sphericase | | Descriptor: | CALCIUM ION, SERINE PROTEASE | | Authors: | Almog, O, Gonzalez, A, Klein, D, Braun, S, Shoham, G. | | Deposit date: | 2001-07-10 | | Release date: | 2002-07-04 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | The 0.93A Crystal Structure of Sphericase: A Calcium-Loaded Serine Protease from Bacillus Sphaericus
J.Mol.Biol., 332, 2003
|
|
2M5P
 
 | |
3V95
 
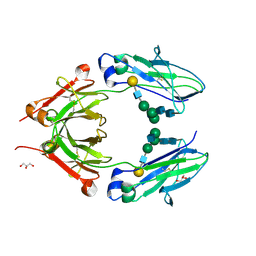 | | Crystal structure of monoclonal human anti-rhesus D Fc and IgG1 t125(yb2/0) in the presence of EDTA | | Descriptor: | GLYCEROL, Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Menez, R, A Stura, E, Bourel, D, Siberil, S, Jorieux, S, De Romeuf, C, Ducancel, F, Fridman, W.H, Teillaud, J.L. | | Deposit date: | 2011-12-23 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Effect of zinc on human IgG1 and its Fc gamma R interactions.
Immunol.Lett., 143, 2012
|
|
3EGS
 
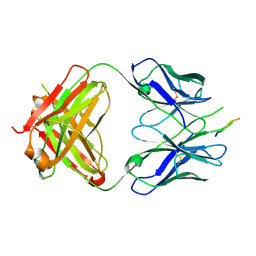 | | Crystal structure of the HIV-1 broadly neutralizing antibody 2F5 in complex with the gp41 scrambledFP-MPER scrHyb3K construct GIGAFGLLGFLAAGSKK-Ahx-K656NEQELLELDKWASLWN671 soaked in ammonium sulfate | | Descriptor: | 2F5 Fab' heavy chain, 2F5 Fab' light chain, gp41 scrFP-MPER construct | | Authors: | Julien, J.-P, Bryson, S, de la Torre, B.G, Andreu, D, Nieva, J.L, Pai, E.F. | | Deposit date: | 2008-09-11 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural constraints imposed by the conserved fusion peptide on the HIV-1 gp41 epitope recognized by the broadly neutralizing antibody 2F5.
J.Phys.Chem.B, 113, 2009
|
|
3GSO
 
 | | Crystal structure of the binary complex between HLA-A2 and HCMV NLV peptide | | Descriptor: | Beta-2-microglobulin, HCMV pp65 fragment 495-503 (NLVPMVATV), HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Saulquin, X, Reiser, J.-B, Debeaupuis, E, Echasserieau, K, Kissenpfennig, A, Legoux, F, Chouquet, A, Le Gorrec, M, Machillot, P, Neveu, B, Thielens, N, Malissen, B, Bonneville, M, Housset, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural bases for the affinity-driven selection of a public TCR against a dominant human cytomegalovirus epitope.
J.Immunol., 183, 2009
|
|
2LVZ
 
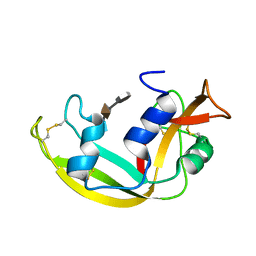 | | Solution structure of a Eosinophil Cationic Protein-trisaccharide heparin mimetic complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-propan-2-yl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, Eosinophil cationic protein | | Authors: | Garcia Mayoral, M, Canales, A, Diaz, D, Lopez Prados, J, Moussaoui, M, de Paz, J, Angulo, J, Nieto, P, Jimenez Barbero, J, Boix, E, Bruix, M. | | Deposit date: | 2012-07-17 | | Release date: | 2013-07-31 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Insights into the glycosaminoglycan-mediated cytotoxic mechanism of eosinophil cationic protein revealed by NMR.
Acs Chem.Biol., 8, 2013
|
|
1EEP
 
 | | 2.4 A RESOLUTION CRYSTAL STRUCTURE OF BORRELIA BURGDORFERI INOSINE 5'-MONPHOSPHATE DEHYDROGENASE IN COMPLEX WITH A SULFATE ION | | Descriptor: | INOSINE 5'-MONOPHOSPHATE DEHYDROGENASE, SULFATE ION | | Authors: | McMillan, F.M, Cahoon, M, White, A, Hedstrom, L, Petsko, G.A, Ringe, D. | | Deposit date: | 2000-02-01 | | Release date: | 2000-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure at 2.4 A resolution of Borrelia burgdorferi inosine 5'-monophosphate dehydrogenase: evidence of a substrate-induced hinged-lid motion by loop 6.
Biochemistry, 39, 2000
|
|
3GTQ
 
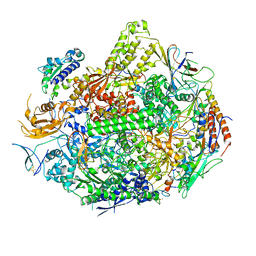 | | Backtracked RNA polymerase II complex induced by damage | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*CP*AP*TP*AP*AP*CP*CP*AP*CP*AP*GP*GP*CP*TP*CP*CP*TP*CP*TP*CP*CP*AP*TP*C)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
1EFW
 
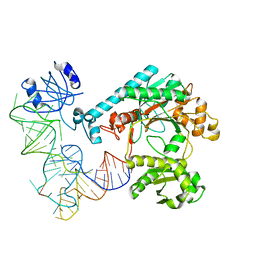 | | Crystal structure of aspartyl-tRNA synthetase from Thermus thermophilus complexed to tRNAasp from Escherichia coli | | Descriptor: | ASPARTYL-TRNA, ASPARTYL-TRNA SYNTHETASE | | Authors: | Briand, C, Poterszman, A, Eiler, S, Webster, G, Thierry, J.-C, Moras, D. | | Deposit date: | 2000-02-10 | | Release date: | 2000-06-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | An intermediate step in the recognition of tRNA(Asp) by aspartyl-tRNA synthetase.
J.Mol.Biol., 299, 2000
|
|
3GW2
 
 | | Crystal structure of possible transcriptional regulatory protein (fragment 1-100) from Mycobacterium bovis. Northeast Structural Genomics Consortium Target MbR242E. | | Descriptor: | Possible transcriptional regulatory arsR-family protein | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-31 | | Release date: | 2009-04-21 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Northeast Structural Genomics Consortium Target MbR242E
To be published
|
|
