7WEK
 
 | |
3VT0
 
 | | Crystal structure of Ct1,3Gal43A in complex with lactose | | Descriptor: | GLYCEROL, Ricin B lectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
5YT6
 
 | |
4MSK
 
 | | Co-crystal structure of tankyrase 1 with compound 34 | | Descriptor: | 3-(4-oxo-3,4-dihydroquinazolin-2-yl)-N-[4-(5-phenyl-1,3,4-oxadiazol-2-yl)phenyl]propanamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2013-09-18 | | Release date: | 2013-12-25 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of novel dual binders as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
3VSZ
 
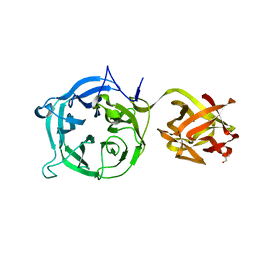 | | Crystal structure of Ct1,3Gal43A in complex with galactan | | Descriptor: | GLYCEROL, Ricin B lectin, beta-D-galactopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
3VSF
 
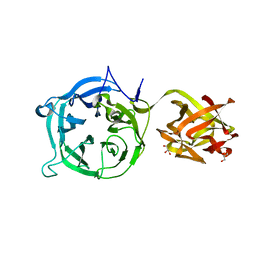 | | Crystal structure of 1,3Gal43A, an exo-beta-1,3-Galactanase from Clostridium thermocellum | | Descriptor: | GLYCEROL, Ricin B lectin | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-04-25 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.757 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
4FM8
 
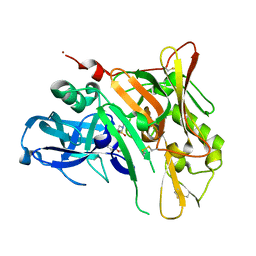 | | Crystal Structure of BACE with Compound 12a | | Descriptor: | (5R,7S)-1-(3-fluorophenyl)-3,7-dimethyl-8-[3-(propan-2-yloxy)benzyl]-2-thia-1,3,8-triazaspiro[4.5]decane 2,2-dioxide, 1,2-ETHANEDIOL, Beta-secretase 1, ... | | Authors: | Vajdos, F.F, Varghese, A.H. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-03 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spirocyclic sulfamides as beta-secretase 1 (BACE-1) inhibitors for the treatment of Alzheimer's disease: utilization of structure based drug design, WaterMap, and CNS penetration studies to identify centrally efficacious inhibitors.
J.Med.Chem., 55, 2012
|
|
5CX3
 
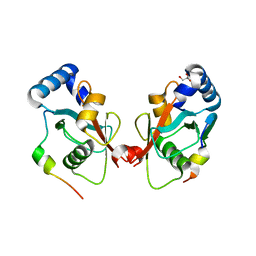 | | Crystal structure of FYCO1 LIR in complex with LC3A | | Descriptor: | FYVE and coiled-coil domain-containing protein 1, GLYCEROL, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Cheng, X, Pan, L. | | Deposit date: | 2015-07-28 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of FYCO1 and MAP1LC3A interaction reveals a novel binding mode for Atg8-family proteins.
Autophagy, 12, 2016
|
|
3OVL
 
 | |
3SZD
 
 | | Crystal structure of Pseudomonas aeruginosa OccK2 (OpdF) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, SULFATE ION, ... | | Authors: | van den Berg, B, Eren, E. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | Substrate Specificity within a Family of Outer Membrane Carboxylate Channels.
Plos Biol., 10, 2012
|
|
3SY9
 
 | |
3T0S
 
 | |
3T35
 
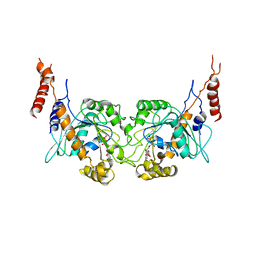 | | Arabidopsis thaliana dynamin-related protein 1A in postfission state | | Descriptor: | Dynamin-related protein 1A, LINKER, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Yan, L.M, Ma, Y.Y, Sun, Y.N, Lou, Z.Y. | | Deposit date: | 2011-07-24 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.592 Å) | | Cite: | Structural basis for mechanochemical role of Arabidopsis thaliana dynamin-related protein in membrane fission
J Mol Cell Biol, 3, 2011
|
|
2ICT
 
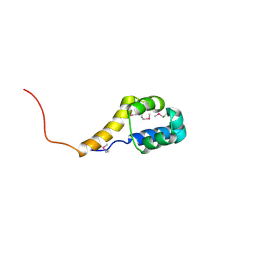 | | Crystal structure of the bacterial antitoxin HigA from Escherichia coli at pH 8.5. Northeast Structural Genomics TARGET ER390. | | Descriptor: | antitoxin higa | | Authors: | Arbing, M.A, Abashidze, M, Hurley, J.M, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Inouye, M, Woychik, N.A, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
3SYB
 
 | |
3T24
 
 | |
2INW
 
 | | Crystal structure of Q83JN9 from Shigella flexneri at high resolution. Northeast Structural Genomics Consortium target SfR137. | | Descriptor: | PHOSPHATE ION, Putative structural protein | | Authors: | Kuzin, A.P, Su, M, Jayaraman, S, Vorobiev, S.M, Wang, D, Jiang, M, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-10-09 | | Release date: | 2006-10-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
3SZV
 
 | |
3VZD
 
 | | Crystal structure of Sphingosine Kinase 1 with inhibitor and ADP | | Descriptor: | 4-{[4-(4-chlorophenyl)-1,3-thiazol-2-yl]amino}phenol, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Min, X, Walker, N.P, Wang, Z. | | Deposit date: | 2012-10-11 | | Release date: | 2013-05-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of sphingosine kinase 1 substrate recognition and catalysis.
Structure, 21, 2013
|
|
2JTD
 
 | |
4PX7
 
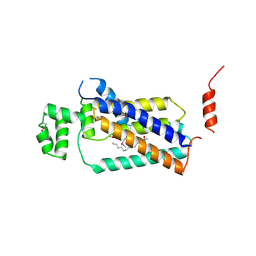 | | Crystal structure of lipid phosphatase E. coli PgpB | | Descriptor: | GLYCEROL, LAURYL DIMETHYLAMINE-N-OXIDE, Phosphatidylglycerophosphatase | | Authors: | Fan, J, Jiang, D, Zhao, Y, Zhang, X.C. | | Deposit date: | 2014-03-22 | | Release date: | 2014-05-28 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of lipid phosphatase Escherichia coli phosphatidylglycerophosphate phosphatase B.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3SY7
 
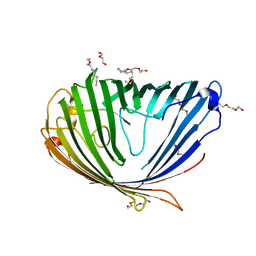 | |
3T20
 
 | |
3SYS
 
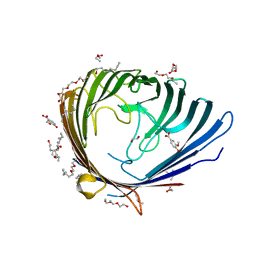 | |
3VZB
 
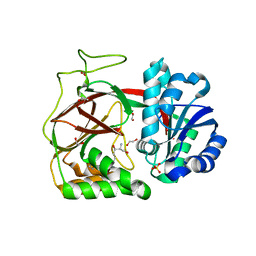 | | Crystal structure of Sphingosine Kinase 1 | | Descriptor: | (2S,3R,4E)-2-aminooctadec-4-ene-1,3-diol, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Min, X, Walker, N.P, Wang, Z. | | Deposit date: | 2012-10-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of sphingosine kinase 1 substrate recognition and catalysis.
Structure, 21, 2013
|
|
