4R8X
 
 | | Crystal structure of a uricase from Bacillus fastidious | | Descriptor: | Uricase | | Authors: | Feng, J, Wang, L, Liu, H.B, Liu, L, Liao, F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Crystal structure of Bacillus fastidious uricase reveals an unexpected folding of the C-terminus residues crucial for thermostability under physiological conditions.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4KY9
 
 | |
6L5Z
 
 | |
8J0H
 
 | |
6PEW
 
 | |
6WHO
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, SODIUM ION, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WHN
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, DI(HYDROXYETHYL)ETHER, Histone deacetylase 2, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WHQ
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, SODIUM ION, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WHZ
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | Histone deacetylase 2, SODIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WI3
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | (SHA)W(DTH)DN(DSN)(DME)(DAS)K peptide macrocycle, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WSJ
 
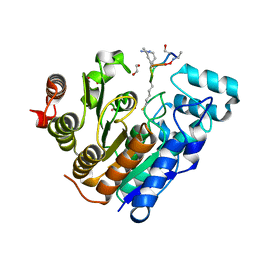 | |
6LKG
 
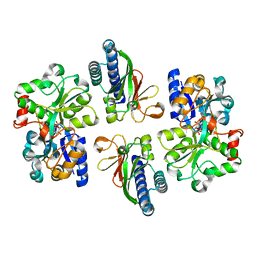 | | two-component system protein mediate signal transduction | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ABC transporter, solute-binding protein, ... | | Authors: | Wang, M, Tao, Y. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Interface switch mediates signal transmission in a two-component system.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PEV
 
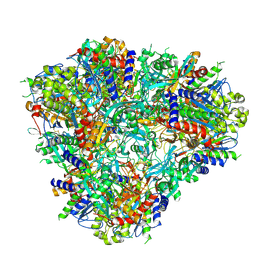 | |
6XBF
 
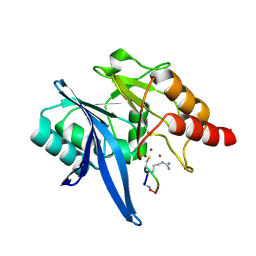 | | Structure of NDM-1 in complex with macrocycle inhibitor NDM1i-1G | | Descriptor: | BlaNDM-4_1_JQ348841, ZINC ION, macrocycle inhibitor NDM1i-1G | | Authors: | Worrall, L.J, Sun, T, Mulligan, V.K, Strynadka, N.C.J. | | Deposit date: | 2020-06-05 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Computationally designed peptide macrocycle inhibitors of New Delhi metallo-beta-lactamase 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6XBE
 
 | | Structure of NDM-1 in complex with macrocycle inhibitor NDM1i-1F | | Descriptor: | BlaNDM-4_1_JQ348841, ZINC ION, macrocycle inhibitor NDM1i-1F | | Authors: | Worrall, L.J, Sun, T, Mulligan, V.K, Strynadka, N.C.J. | | Deposit date: | 2020-06-05 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computationally designed peptide macrocycle inhibitors of New Delhi metallo-beta-lactamase 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6XCI
 
 | | Structure of NDM-1 in complex with macrocycle inhibitor NDM1i-3D | | Descriptor: | ACETATE ION, BlaNDM-4_1_JQ348841, CADMIUM ION, ... | | Authors: | Worrall, L.J, Sun, T, Mulligan, V.K, Strynadka, N.C.J. | | Deposit date: | 2020-06-08 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Computationally designed peptide macrocycle inhibitors of New Delhi metallo-beta-lactamase 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6LKK
 
 | | Two-component system protein mediate signal transduction | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ABC transporter, solute-binding protein | | Authors: | Wang, M, Tao, Y. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Interface switch mediates signal transmission in a two-component system.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8YBC
 
 | |
6PNX
 
 | |
6MWY
 
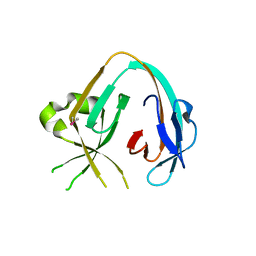 | | The Prp8 intein of Cryptococcus gattii | | Descriptor: | Pre-mRNA-processing-splicing factor 8 | | Authors: | Li, Z, Fu, B, Green, C.M, Lang, Y, Zhang, J, Oven, T.S, Li, X, Callahan, B.P, Chaturvedi, S, Belfort, M, Liao, G, Li, H. | | Deposit date: | 2018-10-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Cisplatin protects mice from challenge ofCryptococcus neoformansby targeting the Prp8 intein.
Emerg Microbes Infect, 8, 2019
|
|
5F22
 
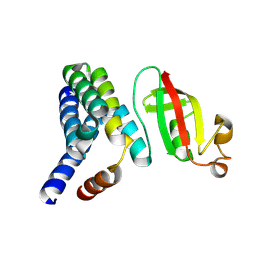 | | C-terminal domain of SARS-CoV nsp8 complex with nsp7 | | Descriptor: | Non-structural protein, Non-structural protein 7 | | Authors: | Li, S. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | C-terminal domain of SARS-CoV nsp8 complex with nsp7
To Be Published
|
|
1PYN
 
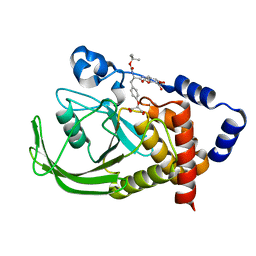 | | DUAL-SITE POTENT, SELECTIVE PROTEIN TYROSINE PHOSPHATASE 1B INHIBITOR USING A LINKED FRAGMENT STRATEGY AND A MALONATE HEAD ON THE FIRST SITE | | Descriptor: | 2-(4-{2-TERT-BUTOXYCARBONYLAMINO-2-[4-(3-HYDROXY-2-METHOXYCARBONYL-PHENOXY)-BUTYLCARBAMOYL]-ETHYL}-PHENOXY)-MALONIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Szczepankiewicz, B.G, Liu, G, Hajduk, P.J, Abad-Zapatero, C, Zhonghua, P, Lubben, T, Trevillyan, J.M, Stashko, M, Ballaron, S.J, Liang, H. | | Deposit date: | 2003-07-09 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and SAR of novel, potent and selective protein tyrosine phosphatase 1B inhibitors.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
7WNQ
 
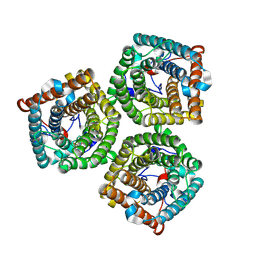 | | Cryo-EM structure of AtSLAC1 S59A mutant | | Descriptor: | Guard cell S-type anion channel SLAC1 | | Authors: | Sun, L, Liu, X, Li, Y. | | Deposit date: | 2022-01-19 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the Arabidopsis guard cell anion channel SLAC1 suggests activation mechanism by phosphorylation.
Nat Commun, 13, 2022
|
|
6WH9
 
 | | Ketoreductase from module 1 of the 6-deoxyerythronolide B synthase (KR1) in complex with antibody fragment (Fab) 1D10 | | Descriptor: | 1D10 (Fab heavy chain), 1D10 (Fab light chain), KR1, ... | | Authors: | Cogan, D.P, Mathews, I.I, Khosla, C. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Antibody Probes of Module 1 of the 6-Deoxyerythronolide B Synthase Reveal an Extended Conformation During Ketoreduction.
J.Am.Chem.Soc., 142, 2020
|
|
8WCK
 
 | | FCP tetramer in Chaetoceros gracilis | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, CHLOROPHYLL A, Chlorophyll a/b-binding protein, ... | | Authors: | Feng, Y, Li, Z, Zhou, C, Shen, J.-R, Liu, C, Wang, W. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural and spectroscopic insights into fucoxanthin chlorophyll a/c-binding proteins of diatoms in diverse oligomeric states.
Plant Commun., 5, 2024
|
|
