8DHV
 
 | | Treponema lecithinolyticum beta-glucuronidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Glycosyl hydrolase family 2, ... | | Authors: | Lietzan, A.D, Redinbo, M.R. | | Deposit date: | 2022-06-28 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Microbial beta-glucuronidases drive human periodontal disease etiology.
Sci Adv, 9, 2023
|
|
8DHE
 
 | |
8DHW
 
 | | Treponema lecithinolyticum beta-glucuronidase in complex with a UNC4917-glucuronide conjugate | | Descriptor: | 4-(4-beta-D-glucopyranuronosylpiperazin-1-yl)-2,7-bis(methylamino)pyrido[3',2':4,5]thieno[3,2-d]pyrimidine, Glycosyl hydrolase family 2, TIM barrel domain protein, ... | | Authors: | Lietzan, A.D, Redinbo, M.R. | | Deposit date: | 2022-06-28 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Microbial beta-glucuronidases drive human periodontal disease etiology.
Sci Adv, 9, 2023
|
|
5XQ1
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | Fermitin family homolog 2,Integrin beta-3 | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8EFN
 
 | | Structure of Sp-STING3 from Stylophora pistillata coral in complex with 3',3'-cGAMP | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Stimulator of interferon genes protein | | Authors: | Li, Y, Slavik, K.M, Morehouse, B.R, Mears, K, Kranzusch, P.J. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8EFM
 
 | | Structure of coral STING receptor from Stylophora pistillata in complex with 2',3'-cGAMP | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein, cGAMP | | Authors: | Li, Y, Slavik, K.M, Morehouse, B.R, Mears, K, Kranzusch, P.J. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
7XP1
 
 | | Crystal structure of PmiR from Pseudomonas aeruginosa | | Descriptor: | ALPHA-METHYLISOCITRIC ACID, GLYCEROL, Probable transcriptional regulator, ... | | Authors: | Zhang, Y.X, Liang, H.H, Gan, J.H. | | Deposit date: | 2022-05-02 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | PmiR senses 2-methylisocitrate levels to regulate bacterial virulence in Pseudomonas aeruginosa.
Sci Adv, 8, 2022
|
|
8E72
 
 | | Treponema lecithinolyticum beta-glucuronidase in complex with a ciprofloxacin-glucuronide conjugate | | Descriptor: | 3-carboxy-1-cyclopropyl-6-fluoro-7-(4-beta-D-glucopyranuronosyl-3,4-dihydropyrazin-1(2H)-yl)-4-oxo-1,4-dihydroquinoline, Glycosyl hydrolase family 2, TIM barrel domain protein, ... | | Authors: | Lietzan, A.D, Redinbo, M.R. | | Deposit date: | 2022-08-23 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Microbial beta-glucuronidases drive human periodontal disease etiology.
Sci Adv, 9, 2023
|
|
8EAY
 
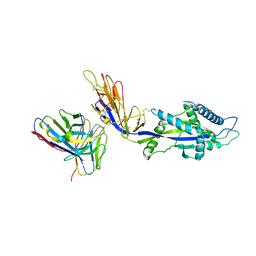 | | HMPV F complex with 4I3 Fab | | Descriptor: | 4I3 heavy chain protein, 4I3 light chain protein, Fusion glycoprotein F0 | | Authors: | Wen, X, Jardetzky, T.S. | | Deposit date: | 2022-08-30 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Potent cross-neutralization of respiratory syncytial virus and human metapneumovirus through a structurally conserved antibody recognition mode.
Cell Host Microbe, 31, 2023
|
|
8E2U
 
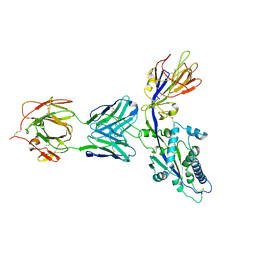 | | HMPV F monomer bound to RSV-199 Fab | | Descriptor: | Fusion glycoprotein F0, RSV-199 heavy chain, RSV-199 light chain | | Authors: | Wen, X, Jardetzky, T.S. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Potent cross-neutralization of respiratory syncytial virus and human metapneumovirus through a structurally conserved antibody recognition mode.
Cell Host Microbe, 31, 2023
|
|
8EBP
 
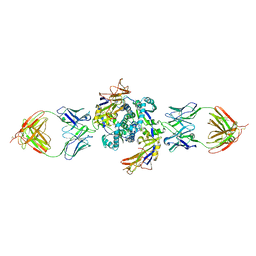 | | HMPV F dimer bound to RSV-199 Fab | | Descriptor: | Fusion glycoprotein F0, RSV-199 heavy chain protein, RSV-199 light chain protein | | Authors: | Wen, X, Jardetzky, T.S. | | Deposit date: | 2022-08-31 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Potent cross-neutralization of respiratory syncytial virus and human metapneumovirus through a structurally conserved antibody recognition mode.
Cell Host Microbe, 31, 2023
|
|
8DZW
 
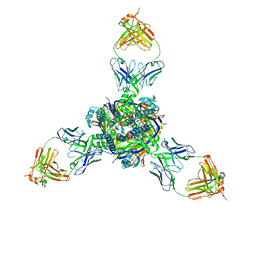 | | RSV F trimer bound to RSV-199 Fab | | Descriptor: | RSV fusion protein, RSV-199 Heavy chain protein, RSV-199 Light chain protein | | Authors: | Wen, X, Jardetzky, T.S. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Potent cross-neutralization of respiratory syncytial virus and human metapneumovirus through a structurally conserved antibody recognition mode.
Cell Host Microbe, 31, 2023
|
|
5HVL
 
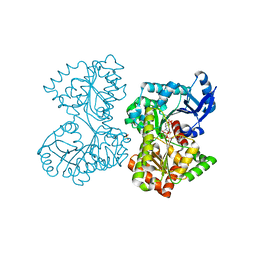 | | Structure of Candida albicans trehalose-6-phosphate synthase in complex with UDP and validoxylamine A | | Descriptor: | (1S,2S,3R,6S)-4-(HYDROXYMETHYL)-6-{[(1S,2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-(HYDROXYMETHYL)CYCLOHEXYL]AMINO}CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha,alpha-trehalose-phosphate synthase [UDP-forming], SULFATE ION, ... | | Authors: | Miao, Y, Brennan, R.G. | | Deposit date: | 2016-01-28 | | Release date: | 2017-05-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structural and In Vivo Studies on Trehalose-6-Phosphate Synthase from Pathogenic Fungi Provide Insights into Its Catalytic Mechanism, Biological Necessity, and Potential for Novel Antifungal Drug Design.
MBio, 8, 2017
|
|
5XPY
 
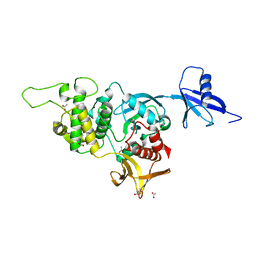 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | ACETATE ION, Fermitin family homolog 2, GLYCEROL | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XPZ
 
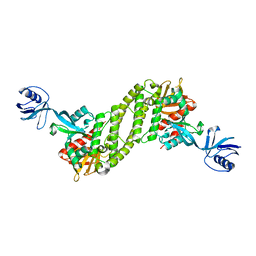 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | Fermitin family homolog 2, GLYCEROL | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5HVO
 
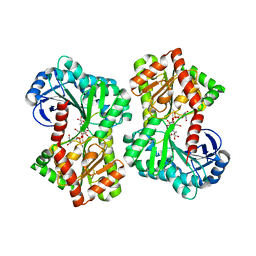 | | Structure of Aspergillus fumigatus trehalose-6-phosphate synthase B in complex with UDP and validoxylamine A | | Descriptor: | (1S,2S,3R,6S)-4-(HYDROXYMETHYL)-6-{[(1S,2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-(HYDROXYMETHYL)CYCLOHEXYL]AMINO}CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha,alpha-trehalose-phosphate synthase (UDP-forming), URIDINE-5'-DIPHOSPHATE | | Authors: | Miao, Y, Brennan, R.G. | | Deposit date: | 2016-01-28 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.467 Å) | | Cite: | Structural and In Vivo Studies on Trehalose-6-Phosphate Synthase from Pathogenic Fungi Provide Insights into Its Catalytic Mechanism, Biological Necessity, and Potential for Novel Antifungal Drug Design.
MBio, 8, 2017
|
|
7DV8
 
 | | The crystal structure of rice immune receptor RGA5-HMA2. | | Descriptor: | Disease resistance protein RGA5 | | Authors: | Zhang, X, Liu, J.F. | | Deposit date: | 2021-01-12 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | A designer rice NLR immune receptor confers resistance to the rice blast fungus carrying noncorresponding avirulence effectors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3VT1
 
 | | Crystal structure of Ct1,3Gal43A in complex with galactose | | Descriptor: | Ricin B lectin, beta-D-galactopyranose | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.187 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
4MSG
 
 | | Crystal structure of tankyrase 1 with compound 22 | | Descriptor: | 3-[(4-oxo-3,4-dihydroquinazolin-2-yl)sulfanyl]-N-[trans-4-(5-phenyl-1,3,4-oxadiazol-2-yl)cyclohexyl]propanamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2013-09-18 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of novel dual binders as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
5CJP
 
 | | The Structural Basis for Cdc42-Induced Dimerization of IQGAPs | | Descriptor: | Cell division cycle 42 (GTP binding protein, 25kDa), isoform CRA_a, ... | | Authors: | Worthylake, D.K, Boyapati, V.K, LeCour Jr, L. | | Deposit date: | 2015-07-14 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structural Basis for Cdc42-Induced Dimerization of IQGAPs.
Structure, 24, 2016
|
|
5XQ0
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | Fermitin family homolog 2,Integrin beta-1, GLYCEROL | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4FM7
 
 | | Crystal Structure of BACE with Compound 14g | | Descriptor: | 4-{[(5R,7S)-1-(3-fluorophenyl)-3,7-dimethyl-2,2-dioxido-2-thia-1,3,8-triazaspiro[4.5]dec-8-yl]methyl}-2-(propan-2-yloxy)phenol, Beta-secretase 1, ZINC ION | | Authors: | Vajdos, F.F, Varghese, A.H. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-03 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Spirocyclic sulfamides as beta-secretase 1 (BACE-1) inhibitors for the treatment of Alzheimer's disease: utilization of structure based drug design, WaterMap, and CNS penetration studies to identify centrally efficacious inhibitors.
J.Med.Chem., 55, 2012
|
|
3VT2
 
 | | Crystal structure of Ct1,3Gal43A in complex with isopropy-beta-D-thiogalactoside | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, GLYCEROL, Ricin B lectin | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
7XP0
 
 | |
5WQ4
 
 | | Crystal structure of OPTN and linear diubiquitin complex | | Descriptor: | Optineurin, ubiquitin | | Authors: | Li, F, Pan, L. | | Deposit date: | 2016-11-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into the ubiquitin recognition by OPTN (optineurin) and its regulation by TBK1-mediated phosphorylation.
Autophagy, 14, 2018
|
|
