5W0I
 
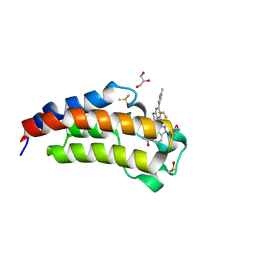 | | CREBBP Bromodomain in complex with Cpd8 (1-(3-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-1-(tetrahydrofuran-3-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | Descriptor: | 1-{3-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-1-[(3S)-oxolan-3-yl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, CREB-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-30 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
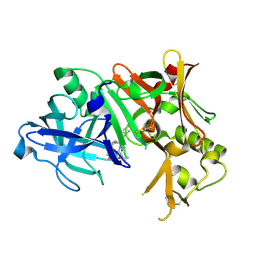
4ZPE
 
 | |
3QNU
 
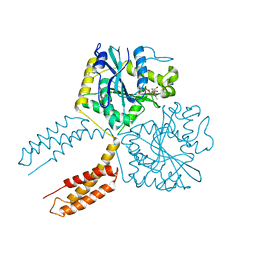 | |
6Y30
 
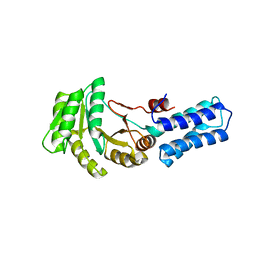 | | NG domain of human SRP54 T115A mutant | | Descriptor: | Signal recognition particle 54 kDa protein | | Authors: | Juaire, K.D, Wild, K, Sinning, I. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and Functional Impact of SRP54 Mutations Causing Severe Congenital Neutropenia.
Structure, 29, 2021
|
|
3QOF
 
 | |
4ZPF
 
 | |
8Q0P
 
 | |
8Q0E
 
 | |
4ZPG
 
 | |
7D0E
 
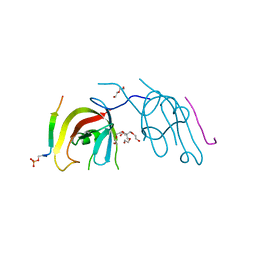 | | Crystal structure of FIP200 Claw/p-CCPG1 FIR2 | | Descriptor: | 3-(2-hydroxyethyloxy)-2-[2-(2-hydroxyethyloxy)ethoxymethyl]-2-(2-hydroxyethyloxymethyl)propan-1-ol, Cell cycle progression protein 1 FIR2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, Z.X, Pan, L.F. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphorylation regulates the binding of autophagy receptors to FIP200 Claw domain for selective autophagy initiation.
Nat Commun, 12, 2021
|
|
6A57
 
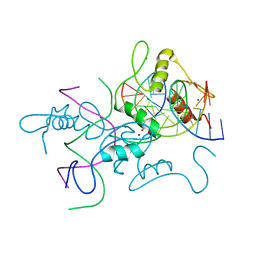 | | Structure of histone demethylase REF6 complexed with DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*TP*CP*TP*CP*TP*GP*TP*TP*TP*TP*GP*TP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*AP*AP*A)-3'), GLYCEROL, ... | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
7CZM
 
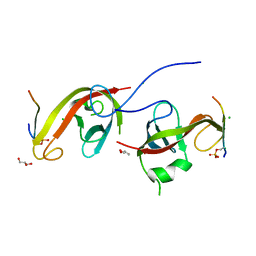 | | Crystal structure of FIP200 Claw/p-OPtineurin LIR complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Optineurin LIR, ... | | Authors: | Zhou, Z.X, Pan, L.F. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylation regulates the binding of autophagy receptors to FIP200 Claw domain for selective autophagy initiation.
Nat Commun, 12, 2021
|
|
7CZG
 
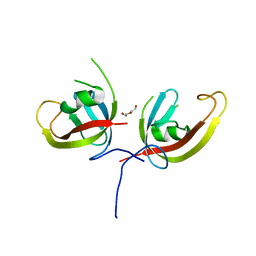 | | Crystal structure of FIP200 Claw domain apo form | | Descriptor: | DI(HYDROXYETHYL)ETHER, RB1-inducible coiled-coil protein 1 | | Authors: | Zhou, Z.X, Pan, L.F. | | Deposit date: | 2020-09-08 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phosphorylation regulates the binding of autophagy receptors to FIP200 Claw domain for selective autophagy initiation.
Nat Commun, 12, 2021
|
|
4JF4
 
 | | OXA-23 meropenem complex | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Smith, C.A, Vakulenko, S.B, Toth, M. | | Deposit date: | 2013-02-27 | | Release date: | 2013-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Basis for Carbapenemase Activity of the OXA-23 beta-Lactamase from Acinetobacter baumannii.
Chem.Biol., 20, 2013
|
|
6YRO
 
 | | Streptococcus suis SadP mutant - N285D | | Descriptor: | GLYCEROL, SODIUM ION, SadP | | Authors: | Papageorgiou, A.C, Haataja, S. | | Deposit date: | 2020-04-20 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The binding mechanism of the virulence factor Streptococcus suis adhesin P subtype to globotetraosylceramide is associated with systemic disease.
J.Biol.Chem., 295, 2020
|
|
5KTU
 
 | | Crystal structure of the bromodomain of human CREBBP bound to pyrazolopiperidine scaffold | | Descriptor: | 1-(3-phenylazanyl-1,4,6,7-tetrahydropyrazolo[4,3-c]pyridin-5-yl)ethanone, CREB-binding protein, DIMETHYL SULFOXIDE | | Authors: | Jayaram, H, Poy, F, Setser, J.W, Bellon, S.F. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Discovery of a Potent and Selective in Vivo Probe (GNE-272) for the Bromodomains of CBP/EP300.
J. Med. Chem., 59, 2016
|
|
7BQ3
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW7647-SRC1 coactivator peptide co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-03-23 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
4JF5
 
 | | Structure of OXA-23 at pH 4.1 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CITRATE ANION | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2013-02-27 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural Basis for Carbapenemase Activity of the OXA-23 beta-Lactamase from Acinetobacter baumannii.
Chem.Biol., 20, 2013
|
|
5ELX
 
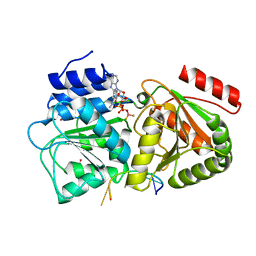 | | S. cerevisiae Dbp5 bound to RNA and mant-ADP BeF3 | | Descriptor: | ATP-dependent RNA helicase DBP5, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Merchant, M.K, Modis, Y. | | Deposit date: | 2015-11-05 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Pi Release Limits the Intrinsic and RNA-Stimulated ATPase Cycles of DEAD-Box Protein 5 (Dbp5).
J.Mol.Biol., 428, 2016
|
|
6NOU
 
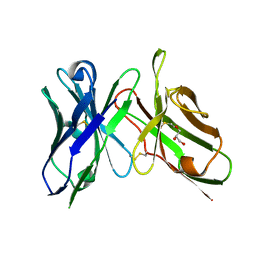 | | An scFv derived from ixekizumab | | Descriptor: | GLYCEROL, scFv derived from ixekizumab | | Authors: | Durbin, J.D, Clawson, D.K, Lu, F, Tian, Y, Lu, J, Atwell, S. | | Deposit date: | 2019-01-16 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Development of tibulizumab, a tetravalent bispecific antibody targeting BAFF and IL-17A for the treatment of autoimmune disease.
Mabs, 11, 2019
|
|
7DHS
 
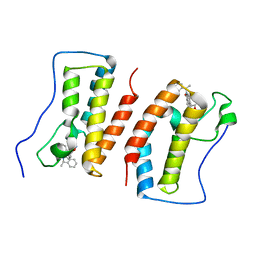 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 6-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1R)-1-phenylethyl]benzo[cd]indol-2-one, Bromodomain-containing protein 4 | | Authors: | Wu, T, Xiang, Q, Wang, C, Wu, C, Zhang, C, Zhang, M, Liu, Z, Zhang, Y, Xiao, L, Xu, Y. | | Deposit date: | 2020-11-17 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Y06014 is a selective BET inhibitor for the treatment of prostate cancer.
Acta Pharmacol.Sin., 42, 2021
|
|
7BQ4
 
 | | X-ray structure of human PPARalpha ligand binding domain-eicosapentaenoic acid (EPA)-SRC1 coactivator peptide co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 5,8,11,14,17-EICOSAPENTAENOIC ACID, GLYCEROL, ... | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-03-23 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
7BPZ
 
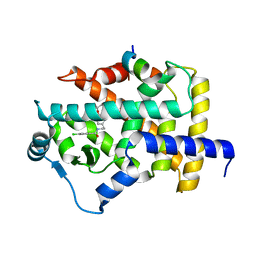 | | X-ray structure of human PPARalpha ligand binding domain-bezafibrate-SRC1 coactivator peptide co-crystals obtained by soaking | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[P-[2-P-CHLOROBENZAMIDO)ETHYL]PHENOXY]-2-METHYLPROPIONIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-03-23 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6A59
 
 | | Structure of histone demethylase REF6 at 1.8A | | Descriptor: | Lysine-specific demethylase REF6, ZINC ION | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
7BQ0
 
 | | X-ray structure of human PPARalpha ligand binding domain-fenofibric acid-SRC1 coactivator peptide co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-03-23 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
