8DD4
 
 | | PI 3-kinase alpha with nanobody 3-142 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1M4B
 
 | | Crystal Structure of Human Interleukin-2 K43C Covalently Modified at C43 with 2-[2-(2-Cyclohexyl-2-guanidino-acetylamino)-acetylamino]-N-(3-mercapto-propyl)-propionamide | | Descriptor: | 2-[2-(2-CYCLOHEXYL-2-GUANIDINO-ACETYLAMINO)-ACETYLAMINO]-N-(3-MERCAPTO-PROPYL)-PROPIONAMIDE, interleukin-2 | | Authors: | Arkin, M.A, Randal, M, DeLano, W.L, Hyde, J, Luong, T.N, Oslob, J.D, Raphael, D.R, Taylor, L, Wang, J, McDowell, R.S, Wells, J.A, Braisted, A.C. | | Deposit date: | 2002-07-02 | | Release date: | 2002-07-31 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Binding of small molecules to an adaptive
protein-protein interface
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
8DD8
 
 | | PI 3-kinase alpha with nanobody 3-142, crosslinked with DSG | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DGY
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 16d (high resolution) | | Descriptor: | 3C-like proteinase, [(1~{R},2~{R})-2-(cyclohexylmethyl)cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, [(1~{R},2~{R})-2-(cyclohexylmethyl)cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-06-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
6GY4
 
 | | Crystal structure of the N-terminal KH domain of human BICC1 | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, CHLORIDE ION, Protein bicaudal C homolog 1, ... | | Authors: | Newman, J.A, Katis, V.L, Shrestha, L, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2018-06-28 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Crystal structure of the N-terminal KH domain of human BICC1
To Be Published
|
|
4A4B
 
 | | Structure of modified phosphoTyr371-c-Cbl-UbcH5B-ZAP-70 complex | | Descriptor: | CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE CBL, TYROSINE-PROTEIN KINASE ZAP-70, ... | | Authors: | Dou, H, Buetow, L, Hock, A, Sibbet, G.J, Vousden, K.H, Huang, D.T. | | Deposit date: | 2011-10-08 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | Structural Basis for Autoinhibition and Phosphorylation-Dependent Activation of C-Cbl
Nat.Struct.Mol.Biol., 19, 2012
|
|
5NSP
 
 | | Crystal structure of TNKS2 in complex with OD334 | | Descriptor: | 1-[4-[4-(2-chlorophenyl)-5-pyrimidin-4-yl-1,2,4-triazol-3-yl]phenyl]-2-oxidanylidene-3~{H}-benzimidazole-5-carbonitrile, BICARBONATE ION, GLYCEROL, ... | | Authors: | Nkizinkiko, Y, Haikarainen, T, Lehtio, L. | | Deposit date: | 2017-04-26 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a Novel Series of Tankyrase Inhibitors by a Hybridization Approach.
J. Med. Chem., 60, 2017
|
|
8SK5
 
 | | Crystal structure of the SARS-CoV-2 neutralizing VHH 7A9 bound to the spike receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, anti-SARS-CoV-2 receptor binding domain VHH | | Authors: | Noland, C.L, Pande, K, Zhang, L, Zhou, H, Galli, J, Eddins, M, Gomez-Llorente, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Discovery and multimerization of cross-reactive single-domain antibodies against SARS-like viruses to enhance potency and address emerging SARS-CoV-2 variants.
Sci Rep, 13, 2023
|
|
6H7G
 
 | | Crystal structure of redox-sensitive phosphoribulokinase (PRK) from the green algae Chlamydomonas reinhardtii | | Descriptor: | Phosphoribulokinase, chloroplastic, SULFATE ION | | Authors: | Fermani, S, Sparla, F, Gurrieri, L, Demitri, N, Polentarutti, M, Falini, G, Trost, P, Lemaire, S.D. | | Deposit date: | 2018-07-31 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | ArabidopsisandChlamydomonasphosphoribulokinase crystal structures complete the redox structural proteome of the Calvin-Benson cycle.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5O4M
 
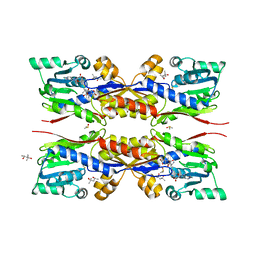 | | Fresh crystals of HcgC from Methanococcus maripaludis cocrystallized with SAH and pyridinol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-carboxy methyl-4-hydroxy-2-pyridinol, DIMETHYL SULFOXIDE, ... | | Authors: | Wagner, T, Bai, L, Xu, T, Hu, X, Ermler, U, Shima, S. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Water-Bridged H-Bonding Network Contributes to the Catalysis of the SAM-Dependent C-Methyltransferase HcgC.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8TBX
 
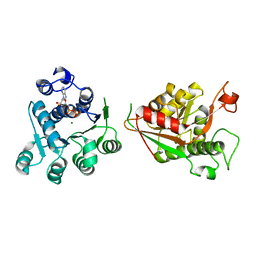 | | Crystal structure of human DDX1 helicase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DDX1, MAGNESIUM ION, ... | | Authors: | Zeng, H, Dong, A, Li, Y, Yen, H, Hejazi, Z, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Halabelian, L. | | Deposit date: | 2023-06-29 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of human DDX1 helicase in complex with ADP
To be published
|
|
8CMJ
 
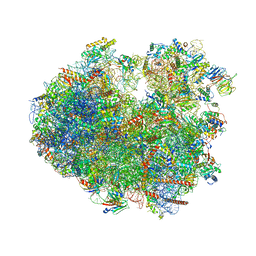 | | Translocation intermediate 4 (TI-4*) of 80S S. cerevisiae ribosome with eEF2 in the absence of sordarin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-02-20 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
8CG8
 
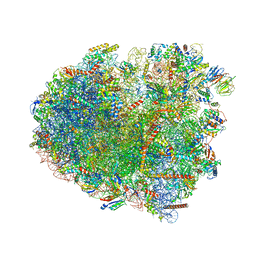 | | Translocation intermediate 3 (TI-3) of 80S S. cerevisiae ribosome with ligands and eEF2 in the presence of sordarin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-02-03 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
5O5R
 
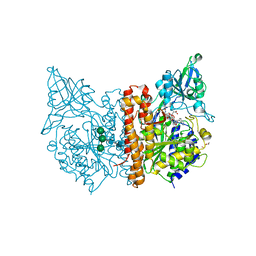 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a urea based inhibitor PSMA 1023 | | Descriptor: | (2~{S})-2-[[(2~{S})-6-[[(2~{S})-2-[[4-[[[(2~{R})-2-[[(2~{R})-2-[(6-fluoranylpyridin-3-yl)carbonylamino]-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]methyl]phenyl]carbonylamino]-3-naphthalen-2-yl-propanoyl]amino]-1-oxidanyl-1-oxidanylidene-hexan-2-yl]carbamoylamino]pentanedioic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Motlova, L, Novakova, Z, Barinka, C. | | Deposit date: | 2017-06-02 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a urea based inhibitor PSMA 1023
To Be Published
|
|
6GI0
 
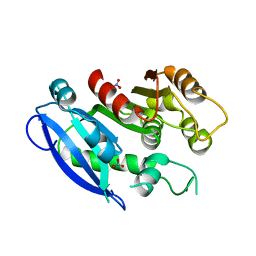 | |
8THV
 
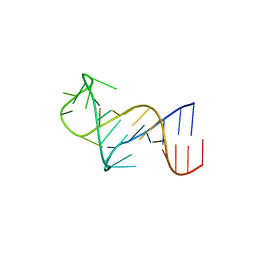 | | FARFAR-NMR ensemble of HIV-1 TAR with apical loop capturing ground and excited conformational states | | Descriptor: | RNA (29-MER) | | Authors: | Roy, R, Geng, A, Shi, H, Merriman, D.K, Dethoff, E.A, Salmon, L, Al-Hashimi, H.M. | | Deposit date: | 2023-07-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Kinetic Resolution of the Atomic 3D Structures Formed by Ground and Excited Conformational States in an RNA Dynamic Ensemble.
J.Am.Chem.Soc., 145, 2023
|
|
8BYJ
 
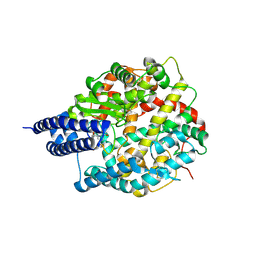 | | The structures of Ace2 in complex with bicyclic peptide inhibitor | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, ALA-CYS-VAL-ARG-SER-HIS-CYS-SER-SER-LEU-LEU-PRO-ARG-ILE-HIS-CYS-ALA, Processed angiotensin-converting enzyme 2, ... | | Authors: | Brear, P, Lulla, A, Harman, M, Dods, R, Chen, L, Bezerra, G, Demydchuk, Y, Stanway, S, Hyvonen, M. | | Deposit date: | 2022-12-13 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Chemical Optimization of Bicyclic Peptide ( Bicycle ) Inhibitors of Angiotensin-Converting Enzyme 2.
J.Med.Chem., 66, 2023
|
|
8CCS
 
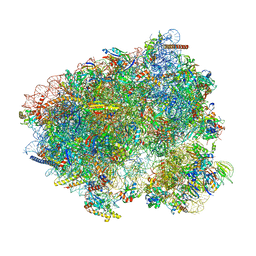 | | 80S S. cerevisiae ribosome with ligands in hybrid-1 pre-translocation (PRE-H1) complex | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-01-27 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (1.97 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
4AA7
 
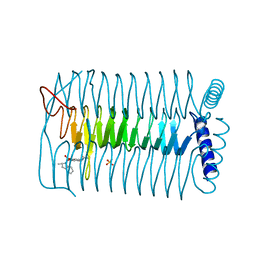 | | E.coli GlmU in complex with an antibacterial inhibitor | | Descriptor: | BIFUNCTIONAL PROTEIN GLMU, N-(2,4-dimethoxy-5-{[(2R)-2-methyl-2,3-dihydro-1H-indol-1-yl]sulfonyl}phenyl)acetamide, SULFATE ION | | Authors: | Otterbein, L, Breed, J, Ogg, D.J. | | Deposit date: | 2011-11-30 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitors of Acetyltransferase Domain of N-Acetylglucosamine-1-Phosphate-Uridyltransferase/ Glucosamine-1-Phosphate-Acetyltransferase (Glmu). Part 1: Hit to Lead Evaluation of a Novel Arylsulfonamide Series.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
8CEH
 
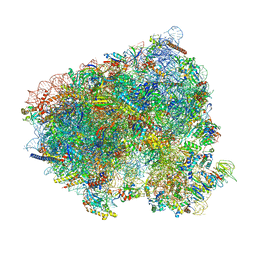 | | Translocation intermediate 4 (TI-4) of 80S S. cerevisiae ribosome with ligands and eEF2 in the presence of sordarin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.05 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
4CK8
 
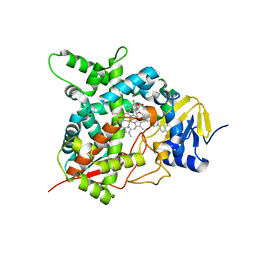 | | STEROL 14-ALPHA DEMETHYLASE (CYP51)FROM TRYPANOSOMA CRUZI IN COMPLEX WITH (R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl 4-(4-(3,4- dichlorophenyl)piperazin-1-yl)phenylcarbamate (LFD) | | Descriptor: | (1R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl {4-[4-(3,4-dichlorophenyl)piperazin-1-yl]phenyl}carbamate, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Friggeri, L, Wawrzak, Z, Tortorella, S, Lepesheva, G.I. | | Deposit date: | 2013-12-30 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural Basis for Rational Design of Inhibitors Targeting Trypanosoma Cruzi Sterol 14Alpha-Demethylase: Two Regions of the Enzyme Molecule Potentiate its Inhibition.
J.Med.Chem., 57, 2014
|
|
6GLV
 
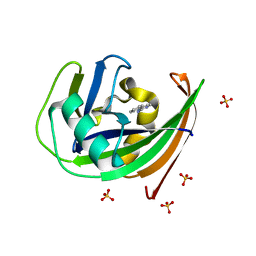 | | Crystal structure of hMTH1 D120N in complex with TH scaffold 1 in the absence of acetate | | Descriptor: | 4-phenylpyrimidin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Eberle, S.A, Wiedmer, L, Sledz, P, Caflisch, A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | hMTH1 D120N in complex with TH scaffold 1
To Be Published
|
|
6GJU
 
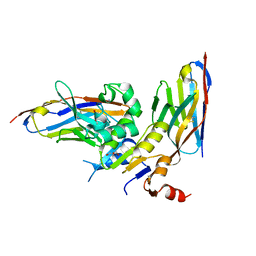 | | human NBD1 of CFTR in complex with nanobodies T2a and T4 | | Descriptor: | Cystic fibrosis transmembrane conductance regulator, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sigoillot, M, Overtus, M, Grodecka, M, Scholl, D, Garcia-Pino, A, Laeremans, T, He, L, Pardon, E, Hildebrandt, E, Urbatsch, I, Steyaert, J, Riordan, J.R, Govaerts, C. | | Deposit date: | 2018-05-17 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Domain-interface dynamics of CFTR revealed by stabilizing nanobodies.
Nat Commun, 10, 2019
|
|
6AGY
 
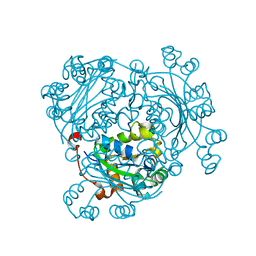 | | Aspergillus fumigatus Af293 NDK | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Hu, Y, Han, L. | | Deposit date: | 2018-08-15 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of crystal structure and key residues of Aspergillus fumigatus nucleoside diphosphate kinase.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
1ME4
 
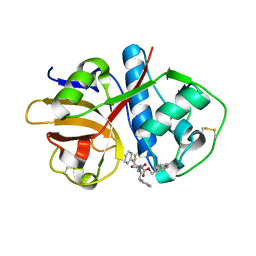 | |
