2RJC
 
 | | Crystal structure of L3MBTL1 protein in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lethal(3)malignant brain tumor-like protein, SULFATE ION | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
5AIZ
 
 | | The PIAS-like coactivator Zmiz1 is a direct and selective cofactor of Notch1 in T-cell development and leukemia | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Cho, H.J, Murai, M, Chiang, M, Cierpicki, T. | | Deposit date: | 2015-02-18 | | Release date: | 2015-10-28 | | Last modified: | 2015-12-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Pias-Like Coactivator Zmiz1 is a Direct and Selective Cofactor of Notch1 in T-Cell Development and Leukemia
Immunity, 43, 2015
|
|
5AC7
 
 | | S. enterica HisA with mutations D7N, D10G, dup13-15 | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, GLYCEROL, SODIUM ION, ... | | Authors: | Newton, M, Guo, X, Soderholm, A, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2015-08-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4WIC
 
 | | Immediate-early 1 protein (IE1) of rhesus macaque cytomegalovirus | | Descriptor: | RhUL123 | | Authors: | Klingl, S, Scherer, M, Sevvana, M, Muller, Y.A, Stamminger, T. | | Deposit date: | 2014-09-25 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Controlled crystal dehydration triggers a space-group switch and shapes the tertiary structure of cytomegalovirus immediate-early 1 (IE1) protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8A3H
 
 | | Cellobiose-derived imidazole complex of the endoglucanase cel5A from Bacillus agaradhaerens at 0.97 A resolution | | Descriptor: | (5R,6R,7R,8S)-7,8-dihydroxy-5-(hydroxymethyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-6-yl beta-D-glucopyranoside, ACETATE ION, GLYCEROL, ... | | Authors: | Varrot, A, Schulein, M, Pipelier, M, Vasella, A, Davies, G.J. | | Deposit date: | 1999-01-20 | | Release date: | 2000-01-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Lateral Protonation of a Glycosidase Inhibitor. Structure of the Bacillus agaradhaerens Cel5A in Complex with a Cellobiose-Derived Imidazole at 0.97 A Resolution
J.Am.Chem.Soc., 121, 1999
|
|
2RJF
 
 | | Crystal structure of L3MBTL1 in complex with H4K20Me2 (residues 12-30), orthorhombic form I | | Descriptor: | Histone H4, Lethal(3)malignant brain tumor-like protein | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
7ZYH
 
 | | Crystal structure of human CPSF30 in complex with hFip1 | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 4, Isoform 4 of Pre-mRNA 3'-end-processing factor FIP1, ZINC ION | | Authors: | Muckenfuss, L.M, Jinek, M, Migenda Herranz, A.C, Clerici, M. | | Deposit date: | 2022-05-24 | | Release date: | 2022-09-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fip1 is a multivalent interaction scaffold for processing factors in human mRNA 3' end biogenesis.
Elife, 11, 2022
|
|
7ZVX
 
 | | Crystal structure of human Annexin A2 in complex with full phosphorothioate 5-10 2'-methoxyethyl DNA gapmer antisense oligonucleotide solved at 2.4 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2'-methoxyethyl DNA gapmer antisense oligonucleotide, Annexin A2, ... | | Authors: | Hyjek-Skladanowska, M, Anderson, B, Mykhaylyk, V, Orr, C, Wagner, A, Skowronek, K, Seth, P, Nowotny, M. | | Deposit date: | 2022-05-17 | | Release date: | 2022-09-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of annexin A2-PS DNA complexes show dominance of hydrophobic interactions in phosphorothioate binding.
Nucleic Acids Res., 51, 2023
|
|
7ZVN
 
 | | Crystal structure of human Annexin A2 in complex with full phosphorothioate 5-10 2'-methoxyethyl DNA gapmer antisense oligonucleotide solved at 1.87 A resolution | | Descriptor: | 2'-methoxyethyl DNA gapmer antisense oligonucleotide, Annexin A2, CALCIUM ION, ... | | Authors: | Hyjek-Skladanowska, M, Anderson, B, Mykhaylyk, V, Orr, C, Wagner, A, Skowronek, K, Seth, P, Nowotny, M. | | Deposit date: | 2022-05-16 | | Release date: | 2022-09-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structures of annexin A2-PS DNA complexes show dominance of hydrophobic interactions in phosphorothioate binding.
Nucleic Acids Res., 51, 2023
|
|
5VRT
 
 | | Nonheme Iron Replacement in a Biosynthetic Nitric Oxide Reductase Model Performing O2 Reduction to Water: Co-bound FeBMb | | Descriptor: | COBALT (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Reed, J, Shi, Y, Zhu, Q, Chakraborty, S, Mirs, E.N, Petrik, I.D, Bhagi-Damodaran, A, Ross, M, Moenne-Loccoz, P, Zhang, Y, Lu, Y. | | Deposit date: | 2017-05-11 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Manganese and Cobalt in the Nonheme-Metal-Binding Site of a Biosynthetic Model of Heme-Copper Oxidase Superfamily Confer Oxidase Activity through Redox-Inactive Mechanism.
J. Am. Chem. Soc., 139, 2017
|
|
1VIH
 
 | | NMR STUDY OF VIGILIN, REPEAT 6, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | VIGILIN | | Authors: | Musco, G, Stier, G, Joseph, C, Morelli, M.A.C, Nilges, M, Gibson, T.J, Pastore, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome.
Cell(Cambridge,Mass.), 85, 1996
|
|
5VVM
 
 | |
1VCZ
 
 | |
5VT8
 
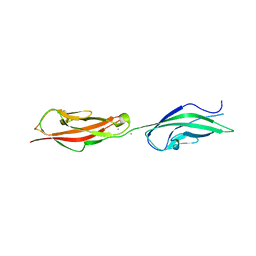 | |
1W18
 
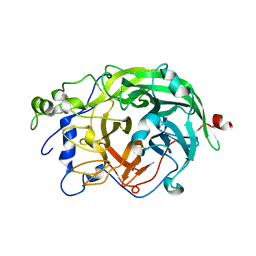 | | Crystal Structure of levansucrase from Gluconacetobacter diazotrophicus | | Descriptor: | LEVANSUCRASE, SULFATE ION | | Authors: | Martinez-Fleites, C, Ortiz-Lombardia, M, Pons, T, Tarbouriech, N, Taylor, E.J, Hernandez, L, Davies, G.J. | | Deposit date: | 2004-06-16 | | Release date: | 2005-05-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Levansucrase from the Gram- Negative Bacterium Gluconacetobacter Diazotrophicus.
Biochem.J., 390, 2005
|
|
1VD8
 
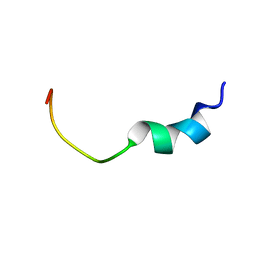 | | Solution structure of FMBP-1 tandem repeat 2 | | Descriptor: | fibroin-modulator-binding-protein-1 | | Authors: | Yamaki, T, Kawaguchi, K, Aizawa, T, Takiya, S, Demura, M, Nitta, K. | | Deposit date: | 2004-03-20 | | Release date: | 2005-03-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FMBP-1 tandem repeat 2
To be Published
|
|
1VE6
 
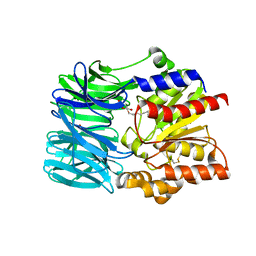 | | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1 | | Descriptor: | Acylamino-acid-releasing enzyme, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Bartlam, M, Wang, G, Gao, R, Yang, H, Zhao, X, Xie, G, Cao, S, Feng, Y, Rao, Z. | | Deposit date: | 2004-03-27 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1
STRUCTURE, 12, 2004
|
|
1WAY
 
 | | Active site thrombin inhibitors | | Descriptor: | 4-[3-(4-CHLOROPHENYL)-1H-PYRAZOL-5-YL]PIPERIDINE, DIMETHYL SULFOXIDE, HIRUGEN, ... | | Authors: | Hartshorn, M.J, Murray, C.W, Cleasby, A, Frederickson, M, Tickle, I.J, Jhoti, H. | | Deposit date: | 2004-10-28 | | Release date: | 2005-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Fragment-Based Lead Discovery Using X-Ray Crystallography
J.Med.Chem., 48, 2005
|
|
5VWI
 
 | |
1VFF
 
 | |
1WBC
 
 | | CRYSTALLIZATION AND PRELIMINARY X-RAY STUDIES OF PSOPHOCARPIN B1, A CHYMOTRYPSIN INHIBITOR FROM WINGED BEAN SEEDS | | Descriptor: | CHYMOTRYPSIN INHIBITOR (WCI) | | Authors: | Dattagupta, J.K, Podder, A, Chakrabarti, C, Sen, U, Dutta, S.K, Singh, M. | | Deposit date: | 1995-11-30 | | Release date: | 1996-04-03 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of a Kunitz-type chymotrypsin from winged bean seeds at 2.95 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1VYI
 
 | | Structure of the c-terminal domain of the polymerase cofactor of rabies virus: insights in function and evolution. | | Descriptor: | GLYCEROL, RNA POLYMERASE ALPHA SUBUNIT | | Authors: | Mavrakis, M, McCarthy, A.A, Roche, S, Blondel, D, Ruigrok, R.W.H. | | Deposit date: | 2004-04-30 | | Release date: | 2004-10-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Function of the C-Terminal Domain of the Polymerase Cofactor of Rabies Virus
J.Mol.Biol., 343, 2004
|
|
5VY5
 
 | | Rabbit muscle aldolase using 200keV | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Herzik Jr, M.A, Wu, M, Lander, G.C. | | Deposit date: | 2017-05-24 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Achieving better-than-3- angstrom resolution by single-particle cryo-EM at 200 keV.
Nat. Methods, 14, 2017
|
|
1VGG
 
 | | Crystal Structure of the Conserved Hypothetical Protein TTHA1091 from Thermus Thermophilus HB8 | | Descriptor: | Conserved Hypothetical Protein TT1634 (TTHA1091) | | Authors: | Satoh, S, Yao, M, Kousumi, Y, Ebihara, A, Matsumoto, K, Okamoto, A, Tanaka, I, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-04-26 | | Release date: | 2004-10-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Conserved Hypothetical Protein TT1634 from Thermus Thermophilus HB8
To be Published
|
|
3D45
 
 | | Crystal structure of mouse PARN in complex with m7GpppG | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Poly(A)-specific ribonuclease PARN | | Authors: | Wu, M, Song, H. | | Deposit date: | 2008-05-13 | | Release date: | 2009-03-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of m(7)GpppG binding to poly(A)-specific ribonuclease.
Structure, 17, 2009
|
|
