6E6J
 
 | | BRD2_Bromodomain2 complex with inhibitor 744 | | Descriptor: | Bromodomain-containing protein 2, N-ethyl-4-[2-(4-fluoro-2,6-dimethylphenoxy)-5-(2-hydroxypropan-2-yl)phenyl]-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide | | Authors: | Longenecker, K.L, Park, C.H, Bigelow, L. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Selective inhibition of the BD2 bromodomain of BET proteins in prostate cancer.
Nature, 578, 2020
|
|
8DYG
 
 | | IL17A homodimer bound to Compound 7 | | Descriptor: | (5P)-2-hydroxy-5-(6-methylquinolin-5-yl)benzoic acid, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
8DYH
 
 | | IL17A homodimer bound to Compound 6 | | Descriptor: | (5P)-N-benzyl-6-chloro-5-(quinolin-5-yl)pyridin-3-amine, GLYCEROL, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
8DYF
 
 | | IL17A homodimer bound to Compound 10 | | Descriptor: | (5M)-3-[({2-[2-(2-{2-[2-({[(5M)-3-carboxy-5-(5,8-dihydroquinolin-4-yl)phenyl]amino}methyl)phenoxy]ethoxy}ethoxy)ethoxy]phenyl}methyl)amino]-5-(quinolin-4-yl)benzoic acid, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
8DYI
 
 | | IL17A homodimer bound to Compound 5 | | Descriptor: | (5P)-5-[5-(benzylamino)pyridin-3-yl]-N-[2-(morpholin-4-yl)ethyl]-1H-indazol-3-amine, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
7RI5
 
 | | Structure of a BAM in MSP1E3D1 nanodiscs at 4 Angstrom resolution | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
7RJ5
 
 | | The structure of BAM in complex with EspP at 7 Angstrom resolution | | Descriptor: | Maltodextrin-binding protein,Autotransporter outer membrane beta-barrel domain-containing protein chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
7RI6
 
 | |
7RI7
 
 | | The structure of BAM in MSP1D1 nanodiscs | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
7RI4
 
 | | Structure of a BAM/EspP(beta9-12) hybrid-barrel intermediate | | Descriptor: | EspPbeta9-12, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
7RI9
 
 | | The structure of BAM in MSP1E3D1 at 6.9 Angstrom resolution | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
7RI8
 
 | | The structure of BAM in MSP2N2 nanodiscs | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
7SIU
 
 | | Crystal structure of HPK1 (MAP4K1) complex with inhibitor A-745 | | Descriptor: | 6-[(5R)-5-benzamidocyclohex-1-en-1-yl]-3-[(1-methyl-1H-pyrazol-4-yl)amino]pyrazine-2-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1, SULFATE ION | | Authors: | Longenecker, K.L, Korepanova, A, Qiu, W. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | The HPK1 Inhibitor A-745 Verifies the Potential of Modulating T Cell Kinase Signaling for Immunotherapy.
Acs Chem.Biol., 17, 2022
|
|
3O2M
 
 | |
3O17
 
 | | Crystal Structure of JNK1-alpha1 isoform | | Descriptor: | C-Jun-amino-terminal kinase-interacting protein 1, JIP1, 10MER PEPTIDE, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2010-07-20 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of JNK1-alpha1 isoform
TO BE PUBLISHED
|
|
6UFR
 
 | | Structure of recombinantly assembled E46K alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein | | Authors: | Eisenberg, D.S, Boyer, D.R, Sawaya, M.R, Li, B, Jiang, L. | | Deposit date: | 2019-09-24 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The alpha-synuclein hereditary mutation E46K unlocks a more stable, pathogenic fibril structure.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5VKC
 
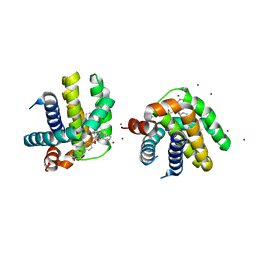 | | Crystal structure of MCL-1 in complex with a BIM competitive inhibitor | | Descriptor: | 7-(3-{[4-(4-acetylpiperazin-1-yl)phenoxy]methyl}-1,5-dimethyl-1H-pyrazol-4-yl)-3-{3-[(naphthalen-1-yl)oxy]propyl}-1-[(pyridin-3-yl)methyl]-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Judge, R.A, Souers, A.J. | | Deposit date: | 2017-04-21 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure-guided design of a series of MCL-1 inhibitors with high affinity and selectivity.
J. Med. Chem., 58, 2015
|
|
5D9Y
 
 | | Crystal structure of TET2-5fC complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*(5FC)P*GP*AP*AP*GP*CP*T)-3'), DNA (5'-D(*AP*GP*CP*TP*TP*CP*GP*AP*CP*AP*GP*T)-3'), FE (III) ION, ... | | Authors: | Hu, L, Cheng, J, Rao, Q, Li, Z, Li, J, Xu, Y. | | Deposit date: | 2015-08-19 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | Structural insight into substrate preference for TET-mediated oxidation.
Nature, 527, 2015
|
|
6B4L
 
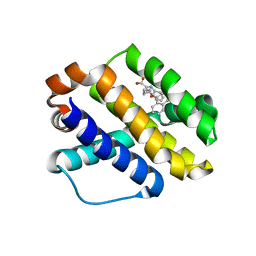 | |
6B4U
 
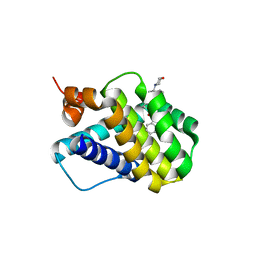 | | Crystal structure of MCL-1 in complex with a BIM competitive inhibitor | | Descriptor: | 7-(2-methylphenyl)-1-[2-(morpholin-4-yl)ethyl]-3-{3-[(naphthalen-1-yl)oxy]propyl}-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Judge, R.A, Souers, A.J. | | Deposit date: | 2017-09-27 | | Release date: | 2017-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design of a series of MCL-1 inhibitors with high affinity and selectivity.
J. Med. Chem., 58, 2015
|
|
7Y9V
 
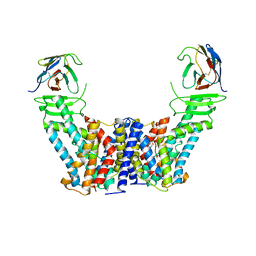 | | Structure of the auxin exporter PIN1 in Arabidopsis thaliana in the IAA-bound state | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Auxin efflux carrier component 1, nanobody | | Authors: | Sun, L, Liu, X, Yang, Z, Xia, J. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into auxin recognition and efflux by Arabidopsis PIN1.
Nature, 609, 2022
|
|
7Y9U
 
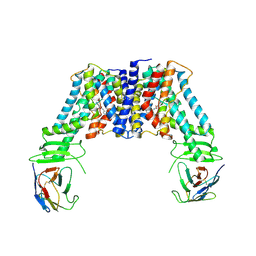 | | Structure of the auxin exporter PIN1 in Arabidopsis thaliana in the NPA-bound state | | Descriptor: | 2-(naphthalen-1-ylcarbamoyl)benzoic acid, Auxin efflux carrier component 1, nanobody | | Authors: | Sun, L, Liu, X, Yang, Z, Xia, J. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into auxin recognition and efflux by Arabidopsis PIN1.
Nature, 609, 2022
|
|
7Y9T
 
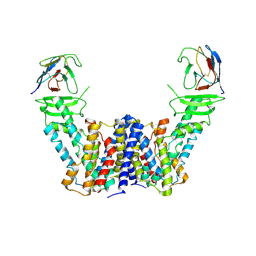 | | Structure of the auxin exporter PIN1 in Arabidopsis thaliana in the apo state | | Descriptor: | Auxin efflux carrier component 1, nanobody | | Authors: | Sun, L, Liu, X, Yang, Z, Xia, J. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into auxin recognition and efflux by Arabidopsis PIN1.
Nature, 609, 2022
|
|
6NCN
 
 | | Fragment-based Discovery of an apoE4 Stabilizer | | Descriptor: | 1-(3-chlorophenyl)cyclobutane-1-carboximidamide, Apolipoprotein E | | Authors: | Jakob, C.G, Qiu, W. | | Deposit date: | 2018-12-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Fragment-Based Discovery of an Apolipoprotein E4 (apoE4) Stabilizer.
J.Med.Chem., 62, 2019
|
|
6NCO
 
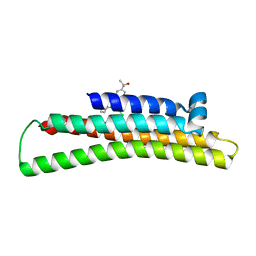 | | Fragment-based Discovery of an apoE4 Stabilizer | | Descriptor: | 1-[5-chloro-4'-(2-hydroxypropan-2-yl)[1,1'-biphenyl]-3-yl]cyclobutane-1-carboximidamide, Apolipoprotein E | | Authors: | Jakob, C.G, Qiu, W. | | Deposit date: | 2018-12-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Fragment-Based Discovery of an Apolipoprotein E4 (apoE4) Stabilizer.
J.Med.Chem., 62, 2019
|
|
