1T9Z
 
 | | Three-dimensional structure of a RNA-polymerase II binding protein. | | Descriptor: | CITRIC ACID, Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION | | Authors: | Kamenski, T, Heilmeier, S, Meinhart, A, Cramer, P. | | Deposit date: | 2004-05-19 | | Release date: | 2004-08-31 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of
RNA Polymerase II CTD Phosphatases.
Mol.Cell, 15, 2004
|
|
1TA0
 
 | | Three-dimensional structure of a RNA-polymerase II binding protein with associated ligand. | | Descriptor: | CITRIC ACID, Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION | | Authors: | Kamenski, T, Heilmeier, S, Meinhart, T, Cramer, P. | | Deposit date: | 2004-05-19 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Mechanism of RNA Polymerase II CTD Phosphatases.
Mol.Cell, 15, 2004
|
|
2PLP
 
 | |
6GXV
 
 | | Amylase in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, A-amylase, CALCIUM ION, ... | | Authors: | Agirre, J, Moroz, O, Meier, S, Brask, J, Munch, A, Hoff, T, Andersen, C, Wilson, K.S, Davies, G.J. | | Deposit date: | 2018-06-27 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The structure of the AliC GH13 alpha-amylase from Alicyclobacillus sp. reveals the accommodation of starch branching points in the alpha-amylase family.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6GYA
 
 | | Amylase in complex with branched ligand | | Descriptor: | A-amylase, CALCIUM ION, SODIUM ION, ... | | Authors: | Agirre, J, Moroz, O, Meier, S, Brask, J, Munch, A, Hoff, T, Andersen, C, Wilson, K.S, Davies, G.J. | | Deposit date: | 2018-06-28 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The structure of the AliC GH13 alpha-amylase from Alicyclobacillus sp. reveals the accommodation of starch branching points in the alpha-amylase family.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5OOW
 
 | | Crystal structure of lobe II from the nucleotide binding domain of DnaK in complex with AMPPCP | | Descriptor: | Chaperone protein DnaK, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Jakob, R.P, Bauer, D, Meinhold, S, Stigler, J, Merkel, U, Maier, T, Rief, M, Zoldak, G. | | Deposit date: | 2017-08-09 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A folding nucleus and minimal ATP binding domain of Hsp70 identified by single-molecule force spectroscopy.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7S0I
 
 | | CRYSTAL STRUCTURE OF N1 NEURAMINIDASE FROM A/Michigan/45/2015(H1N1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2021-08-30 | | Release date: | 2021-12-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.892 Å) | | Cite: | A Novel Recombinant Influenza Virus Neuraminidase Vaccine Candidate Stabilized by a Measles Virus Phosphoprotein Tetramerization Domain Provides Robust Protection from Virus Challenge in the Mouse Model.
Mbio, 12, 2021
|
|
5ZUH
 
 | |
7F61
 
 | |
7T3D
 
 | | CryoEM map of anchor 222-1C06 Fab and lateral patch 2B05 Fab binding H1 HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 222-1C06 mAb heavy chain, ... | | Authors: | Han, J, Ward, A.B. | | Deposit date: | 2021-12-07 | | Release date: | 2022-01-12 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Broadly neutralizing antibodies target a haemagglutinin anchor epitope.
Nature, 602, 2022
|
|
6TFI
 
 | | PXR IN COMPLEX WITH THROMBIN INHIBITOR COMPOUND 17 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Hillig, R.C, Puetter, V. | | Deposit date: | 2019-11-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design, Synthesis, and Pharmacological Characterization of a Neutral, Non-Prodrug Thrombin Inhibitor with Good Oral Pharmacokinetics.
J.Med.Chem., 63, 2020
|
|
7OAE
 
 | | Cryo-EM structure of the plectasin fibril (double strands) | | Descriptor: | Fungal defensin plectasin | | Authors: | Effantin, G. | | Deposit date: | 2021-04-19 | | Release date: | 2022-04-27 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | pH- and concentration-dependent supramolecular assembly of a fungal defensin plectasin variant into helical non-amyloid fibrils.
Nat Commun, 13, 2022
|
|
7OAG
 
 | | Cryo-EM structure of the plectasin fibril (single strand) | | Descriptor: | Fungal defensin plectasin | | Authors: | Effantin, G. | | Deposit date: | 2021-04-19 | | Release date: | 2022-04-27 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | pH- and concentration-dependent supramolecular assembly of a fungal defensin plectasin variant into helical non-amyloid fibrils.
Nat Commun, 13, 2022
|
|
5LRA
 
 | | Plastidial phosphorylase PhoI from barley in complex with maltotetraose | | Descriptor: | Alpha-1,4 glucan phosphorylase, PYRIDOXAL-5'-PHOSPHATE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Cuesta-Seijo, J.A, Ruzanski, C, Krucewicz, K, Palcic, M.M. | | Deposit date: | 2016-08-18 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional and structural characterization of plastidic starch phosphorylase during barley endosperm development.
PLoS ONE, 12, 2017
|
|
7A43
 
 | |
7A42
 
 | |
7A45
 
 | | CO-bound sperm whale myoglobin measured by serial femtosecond crystallography | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mehrabi, P, Schulz, E.C, Buecker, R. | | Deposit date: | 2020-08-19 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Serial femtosecond and serial synchrotron crystallography can yield data of equivalent quality: A systematic comparison.
Sci Adv, 7, 2021
|
|
7A44
 
 | | CO-bound sperm whale myoglobin measured by serial synchrotron crystallography | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mehrabi, P, Schulz, E.C, Buecker, R. | | Deposit date: | 2020-08-19 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Serial femtosecond and serial synchrotron crystallography can yield data of equivalent quality: A systematic comparison.
Sci Adv, 7, 2021
|
|
8B69
 
 | | Heterotetramer of K-Ras4B(G12V) and Rgl2(RBD) | | Descriptor: | Isoform 2B of GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Tariq, M, Fairall, L, Romartinez-Alonso, B, Dominguez, C, Schwabe, J.W.R, Tanaka, K. | | Deposit date: | 2022-09-26 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structural insights into the complex of oncogenic KRas4B G12V and Rgl2, a RalA/B activator.
Life Sci Alliance, 7, 2024
|
|
5LBY
 
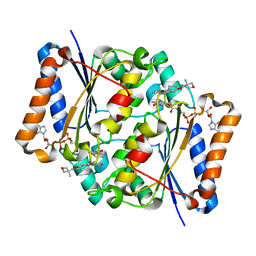 | | Structure of the human quinone reductase 2 (NQO2) in complex with crenolanib | | Descriptor: | 1-(2-{5-[(3-Methyloxetan-3-yl)methoxy]-1H-benzimidazol-1-yl}quinolin-8-yl)piperidin-4-amine, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Schneider, S, Medard, G, Kuester, B. | | Deposit date: | 2016-06-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
5LBW
 
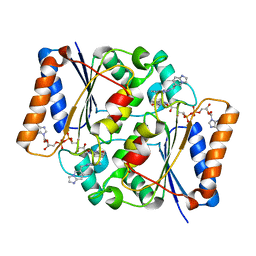 | | Structure of the human quinone reductase 2 (NQO2) in complex with volitinib | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ZINC ION, ... | | Authors: | Schneider, S, Medard, G, Kuester, B. | | Deposit date: | 2016-06-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
5LBZ
 
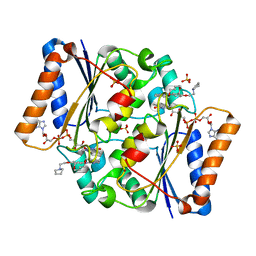 | | Structure of the human quinone reductase 2 (NQO2) in complex with pacritinib | | Descriptor: | 11-(2-pyrrolidin-1-yl-ethoxy)-14,19-dioxa-5,7,26-triaza-tetracyclo[19.3.1.1(2,6).1(8,12)]heptacosa-1(25),2(26),3,5,8,10,12(27),16,21,23-decaene, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Schneider, S, Medard, G, Kuster, B. | | Deposit date: | 2016-06-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
7AQE
 
 | | Structure of SARS-CoV-2 Main Protease bound to UNC-2327 | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Meents, A. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6ZUG
 
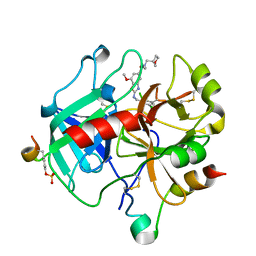 | | Crystal Structure of Thrombin in complex with compound10 | | Descriptor: | 2-[(3-chlorophenyl)methylamino]-7-methoxy-~{N}-[[(3~{S})-oxolan-3-yl]methyl]-~{N}-propyl-1,3-benzoxazole-5-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin-2, ... | | Authors: | Schafer, M. | | Deposit date: | 2020-07-22 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Pharmacological Characterization of a Neutral, Non-Prodrug Thrombin Inhibitor with Good Oral Pharmacokinetics.
J.Med.Chem., 63, 2020
|
|
6ZUU
 
 | | Crystal structure of Thrombin in complex with compound30 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin-2, Prothrombin, ... | | Authors: | Schafer, M. | | Deposit date: | 2020-07-23 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Design, Synthesis, and Pharmacological Characterization of a Neutral, Non-Prodrug Thrombin Inhibitor with Good Oral Pharmacokinetics.
J.Med.Chem., 63, 2020
|
|
