2FHW
 
 | | Solution structure of human relaxin-3 | | Descriptor: | Relaxin 3 (Prorelaxin H3) (Insulin-like peptide INSL7) (Insulin-like peptide 7) | | Authors: | Rosengren, K.J, Craik, D.J. | | Deposit date: | 2005-12-27 | | Release date: | 2006-01-24 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure and novel insights into the determinants of the receptor specificity of human relaxin-3.
J.Biol.Chem., 281, 2006
|
|
2H8B
 
 | | Solution structure of INSL3 | | Descriptor: | Insulin-like 3 | | Authors: | Rosengren, K.J, Craik, D.J, Daly, N.L. | | Deposit date: | 2006-06-07 | | Release date: | 2006-08-01 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Characterization of the LGR8 Receptor Binding Surface of Insulin-like Peptide 3
J.Biol.Chem., 281, 2006
|
|
7O4W
 
 | |
7P40
 
 | | P5C3 is a potent fab neutralizer | | Descriptor: | Spike glycoprotein, Variable Heavy Chain P5C3 (VH), Variable Light Chain P5C3 (VL) | | Authors: | perez, L. | | Deposit date: | 2021-07-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A highly potent antibody effective against SARS-CoV-2 variants of concern.
Cell Rep, 37, 2021
|
|
7PHG
 
 | | MaP OF P5C3RBD Interface | | Descriptor: | Heavy ChaIn variable, Light ChaIn, Surface glycoprotein | | Authors: | Perez, L. | | Deposit date: | 2021-08-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A highly potent antibody effective against SARS-CoV-2 variants of concern.
Cell Rep, 37, 2021
|
|
2X5I
 
 | | Crystal structure echovirus 7 | | Descriptor: | LAURIC ACID, VP1, VP2, ... | | Authors: | Plevka, P, Hafenstein, S, Zhang, Y, Bowman, V.D, Chipman, P.R, Bator, C.M, Rossmann, M.G. | | Deposit date: | 2010-02-08 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Interaction of Decay-Accelerating Factor with Echovirus 7.
J.Virol., 84, 2010
|
|
7MBI
 
 | | Structure of SARS-CoV2 3CL protease covalently bound to peptidomimetic inhibitor | | Descriptor: | 2,4,6-trimethylpyridine-3-carboxylic acid, 3C-like proteinase, 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-3-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide | | Authors: | Khan, M.B, Lu, J, Young, H.S, Lemieux, M.J. | | Deposit date: | 2021-03-31 | | Release date: | 2021-07-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Peptidomimetic alpha-Acyloxymethylketone Warheads with Six-Membered Lactam P1 Glutamine Mimic: SARS-CoV-2 3CL Protease Inhibition, Coronavirus Antiviral Activity, and in Vitro Biological Stability.
J.Med.Chem., 65, 2022
|
|
7OSE
 
 | | cytochrome bd-II type oxidase with bound aurachin D | | Descriptor: | Aurachin D, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome bd-II ubiquinol oxidase subunit 1, ... | | Authors: | Grauel, A, Kaegi, J, Rasmussen, T, Wohlwend, D, Boettcher, B, Friedrich, T. | | Deposit date: | 2021-06-08 | | Release date: | 2021-11-17 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of Escherichia coli cytochrome bd-II type oxidase with bound aurachin D.
Nat Commun, 12, 2021
|
|
8P5W
 
 | | Single particle cryo-EM structure of homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum following reaction with the 2-oxoglutarate analogue succinyl phosphonate | | Descriptor: | (4~{S})-4-[(2~{R})-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-2~{H}-1,3-thiazol-2-yl]-4-oxidanyl-4-phosphono-butanoic acid, 2-oxoglutarate dehydrogenase E1/E2 component, ACETYL COENZYME *A, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
5SVO
 
 | | Structure of IDH2 mutant R140Q | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Isocitrate dehydrogenase [NADP], mitochondrial, ... | | Authors: | Xie, X, Kulathila, R. | | Deposit date: | 2016-08-06 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Allosteric Mutant IDH1 Inhibitors Reveal Mechanisms for IDH1 Mutant and Isoform Selectivity.
Structure, 25, 2017
|
|
5SVN
 
 | | Structure of IDH2 mutant R172K | | Descriptor: | DI(HYDROXYETHYL)ETHER, Isocitrate dehydrogenase [NADP], mitochondrial, ... | | Authors: | Xie, X, Kulathila, R. | | Deposit date: | 2016-08-06 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Allosteric Mutant IDH1 Inhibitors Reveal Mechanisms for IDH1 Mutant and Isoform Selectivity.
Structure, 25, 2017
|
|
8P5S
 
 | | Crystal structure of the homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYL COENZYME *A, ... | | Authors: | Yang, L, Boyko, A, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.459 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8P5T
 
 | | Single particle cryo-EM structure of the homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, ACETYL COENZYME *A, MAGNESIUM ION, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8P5R
 
 | | Crystal structure of full-length, homohexameric 2-oxoglutarate dehydrogenase KGD from Mycobacterium smegmatis in complex with GarA | | Descriptor: | CALCIUM ION, Glycogen accumulation regulator GarA, MAGNESIUM ION, ... | | Authors: | Wagner, T, Mechaly, A.M, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.562 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8P5V
 
 | | Single particle cryo-EM structure of homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum in complex with the product succinyl-CoA | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, ACETYL COENZYME *A, MAGNESIUM ION, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.07 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8P5U
 
 | | Single particle cryo-EM structure of homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum with Coenzyme A bound to the E2o domain | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, ACETYL COENZYME *A, COENZYME A, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8P5X
 
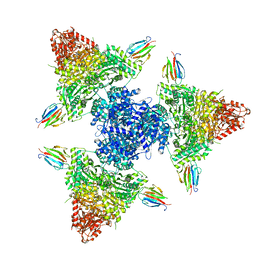 | | Single particle cryo-EM structure of the complex between Corynebacterium glutamicum homohexameric 2-oxoglutarate dehydrogenase OdhA and the FHA-protein inhibitor OdhI | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, MAGNESIUM ION, Oxoglutarate dehydrogenase inhibitor, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
5SUN
 
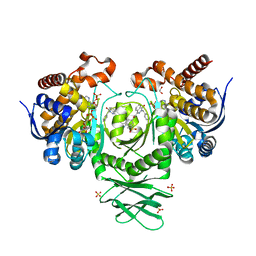 | | IDH1 R132H in complex with IDH146 | | Descriptor: | 1,2-ETHANEDIOL, 3-benzyl-N-[3-(dimethylsulfamoyl)phenyl]-4-oxo-3,4-dihydrophthalazine-1-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Xie, X, Kulathila, R. | | Deposit date: | 2016-08-03 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.477 Å) | | Cite: | Allosteric Mutant IDH1 Inhibitors Reveal Mechanisms for IDH1 Mutant and Isoform Selectivity.
Structure, 25, 2017
|
|
5SVF
 
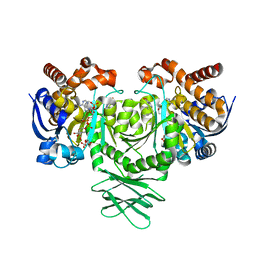 | | IDH1 R132H in complex with IDH125 | | Descriptor: | (4S)-3-(2-{[(1S)-1-phenylethyl]amino}pyrimidin-4-yl)-4-(propan-2-yl)-1,3-oxazolidin-2-one, CITRATE ANION, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Xie, X, Kulathila, R. | | Deposit date: | 2016-08-05 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Allosteric Mutant IDH1 Inhibitors Reveal Mechanisms for IDH1 Mutant and Isoform Selectivity.
Structure, 25, 2017
|
|
5TQH
 
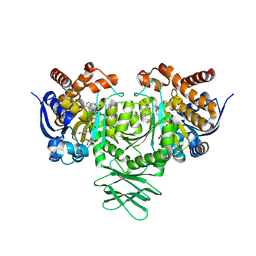 | | IDH1 R132H mutant in complex with IDH889 | | Descriptor: | (4S)-3-[2-({(1S)-1-[5-(4-fluoro-3-methylphenyl)pyrimidin-2-yl]ethyl}amino)pyrimidin-4-yl]-4-(propan-2-yl)-1,3-oxazolidin-2-one, CITRATE ANION, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Xie, X, Kulathila, R. | | Deposit date: | 2016-10-24 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of 3-Pyrimidin-4-yl-oxazolidin-2-ones as Allosteric and Mutant Specific Inhibitors of IDH1.
ACS Med Chem Lett, 8, 2017
|
|
8PQ2
 
 | | XBB 1.0 RBD bound to P4J15 (Local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, P4J15 Fragment Antigen-Binding Heavy Chain, P4J15 Fragment Antigen-Binding Light Chain, ... | | Authors: | Duhoo, Y, Lau, K. | | Deposit date: | 2023-07-10 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Broadly potent anti-SARS-CoV-2 antibody shares 93% of epitope with ACE2 and provides full protection in monkeys.
J Infect, 87, 2023
|
|
8PSD
 
 | | SARS-CoV-2 XBB 1.0 closed conformation. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Duhoo, Y, Lau, K. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Broadly potent anti-SARS-CoV-2 antibody shares 93% of epitope with ACE2 and provides full protection in monkeys.
J Infect, 87, 2023
|
|
5KKG
 
 | | Crystal structure of E72A mutant of ancestral protein ancMT of ADP-dependent sugar kinases family | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, IODIDE ION, ... | | Authors: | Castro-Fernandez, V, Herrera-Morande, A, Zamora, R, Merino, F, Pereira, H.M, Brandao-Neto, J, Garratt, R, Guixe, V. | | Deposit date: | 2016-06-21 | | Release date: | 2017-07-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | Reconstructed ancestral enzymes reveal that negative selection drove the evolution of substrate specificity in ADP-dependent kinases.
J. Biol. Chem., 292, 2017
|
|
6G5J
 
 | | Secreted phospholipase A2 type X in complex with ligand | | Descriptor: | (3~{R})-3-[3-[2-aminocarbonyl-6-(trifluoromethyloxy)indol-1-yl]phenyl]butanoic acid, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sandmark, J, Oster, L. | | Deposit date: | 2018-03-29 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design of Selective sPLA2-X Inhibitor (-)-2-{2-[Carbamoyl-6-(trifluoromethoxy)-1H-indol-1-yl]pyridine-2-yl}propanoic Acid.
ACS Med Chem Lett, 9, 2018
|
|
6QAY
 
 | |
