1CEF
 
 | | CEFOTAXIME COMPLEXED WITH THE STREPTOMYCES R61 DD-PEPTIDASE | | Descriptor: | CEFOTAXIME, C3' cleaved, open, ... | | Authors: | Knox, J.R, Kuzin, A.P. | | Deposit date: | 1995-01-12 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Binding of cephalothin and cefotaxime to D-ala-D-ala-peptidase reveals a functional basis of a natural mutation in a low-affinity penicillin-binding protein and in extended-spectrum beta-lactamases.
Biochemistry, 34, 1995
|
|
1CEG
 
 | | CEPHALOTHIN COMPLEXED WITH DD-PEPTIDASE | | Descriptor: | CEPHALOTHIN GROUP, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE TRANSPEPTIDASE | | Authors: | Knox, J.R, Kuzin, A.P. | | Deposit date: | 1995-01-12 | | Release date: | 1996-10-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of cephalothin and cefotaxime to D-ala-D-ala-peptidase reveals a functional basis of a natural mutation in a low-affinity penicillin-binding protein and in extended-spectrum beta-lactamases.
Biochemistry, 34, 1995
|
|
4JCA
 
 | | Crystal Structure of the apo form of the evolved variant of the computationally designed serine hydrolase, OSH55.4_H1. Northeast Structural Genomics Consortium (NESG) Target OR273 | | Descriptor: | CITRIC ACID, RUBIDIUM ION, serine hydrolase | | Authors: | Kuzin, A.P, Lew, S, Rajagopalan, S, Seetharaman, J, Tong, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-02-21 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
1TVG
 
 | | X-ray structure of human PP25 gene product, HSPC034. Northeast Structural Genomics Target HR1958. | | Descriptor: | CALCIUM ION, LOC51668 protein, SAMARIUM (III) ION | | Authors: | Kuzin, A.P, Vorobiev, S.M, Lee, I, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-29 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study.
Proteins, 75, 2009
|
|
1SDJ
 
 | | X-RAY STRUCTURE OF YDDE_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ET25. | | Descriptor: | Hypothetical protein yddE, SULFATE ION | | Authors: | Kuzin, A.P, Edstrom, W, Skarina, T, Korniyenko, Y, Savchenko, A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-02-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
2INW
 
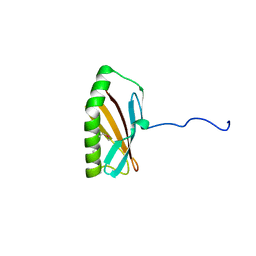 | | Crystal structure of Q83JN9 from Shigella flexneri at high resolution. Northeast Structural Genomics Consortium target SfR137. | | Descriptor: | PHOSPHATE ION, Putative structural protein | | Authors: | Kuzin, A.P, Su, M, Jayaraman, S, Vorobiev, S.M, Wang, D, Jiang, M, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-10-09 | | Release date: | 2006-10-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
2G2X
 
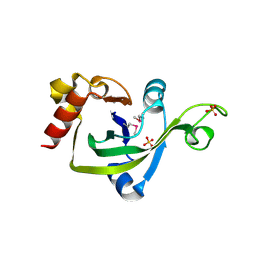 | | X-Ray Crystal Structure Protein Q88CH6 from Pseudomonas putida. Northeast Structural Genomics Consortium Target PpR72. | | Descriptor: | SULFATE ION, hypothetical protein PP5205 | | Authors: | Kuzin, A.P, Chen, Y, Abashidze, M, Acton, T, Conover, K, Janjua, H, Ma, L.-C, Ho, C.K, Cunningham, K, Montelione, G, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-02-16 | | Release date: | 2006-03-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural genomics reveals EVE as a new ASCH/PUA-related domain.
Proteins, 75, 2009
|
|
1ACX
 
 | |
3KH2
 
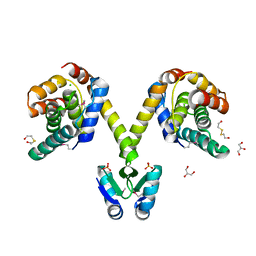 | | Crystal structure of the P1 bacteriophage Doc toxin (F68S) in complex with the Phd antitoxin (L17M/V39A). Northeast Structural Genomics targets ER385-ER386 | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Death on curing protein, ... | | Authors: | Arbing, M.A, Kuzin, A.P, Su, M, Abashidze, M, Verdon, G, Liu, M, Xiao, R, Acton, T, Inouye, M, Montelione, G.T, Woychik, N.A, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-29 | | Release date: | 2010-08-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
3B55
 
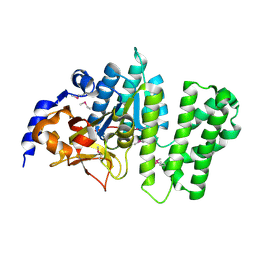 | | Crystal structure of the Q81BN2_BACCR protein from Bacillus cereus. NESG target BcR135 | | Descriptor: | CALCIUM ION, Succinoglycan biosynthesis protein | | Authors: | Vorobiev, S.M, Chen, Y, Kuzin, A.P, Seetharaman, J, Wang, H, Cunningham, K, Owens, L, Maglaqui, M, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-10-25 | | Release date: | 2007-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Q81BN2_BACCR protein from Bacillus cereus.
To be Published
|
|
2A7W
 
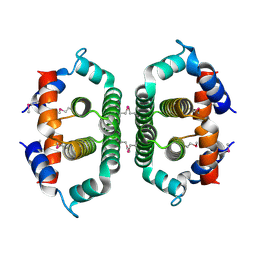 | | Crystal Structure of Phosphoribosyl-ATP Pyrophosphatase from Chromobacterium violaceum (ATCC 12472). NESG TARGET CVR7 | | Descriptor: | Phosphoribosyl-ATP pyrophosphatase | | Authors: | Benach, J, Forouhar, F, Kuzin, A.P, Abashidze, M, Vorobiev, S.M, Rong, X, Acton, T.B, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-07-06 | | Release date: | 2005-10-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Phosphoribosyl-ATP Pyrophosphatase from Chromobacterium violaceum. NESG Target CVR7.
To be Published
|
|
2AEG
 
 | | X-Ray Crystal Structure of Protein Atu5096 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR63. | | Descriptor: | hypothetical protein AGR_pAT_140 | | Authors: | Forouhar, F, Abashidze, M, Kuzin, A.P, Vorobiev, S.M, Shastry, R, Cooper, B, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-07-22 | | Release date: | 2005-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Hypothetical Protein Atu5096 from Agrobacterium tumefaciens.
To be Published
|
|
2AP6
 
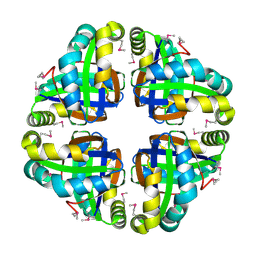 | | X-Ray Crystal Structure of Protein Atu4242 from Agrobacterium tumefaciens. Northeast Strucutral Genomics Consortium Target AtR43. | | Descriptor: | hypothetical protein Atu4242 | | Authors: | Benach, J, Kuzin, A.P, Forouhar, F, Abashidze, M, Vorobiev, S.M, Rong, X, Acton, T.B, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a Hypothetical protein Atu4242 from Agrobacterium tumefaciens (strain C58 / ATCC 3 NESG Target ATR43.
To be Published
|
|
1FSG
 
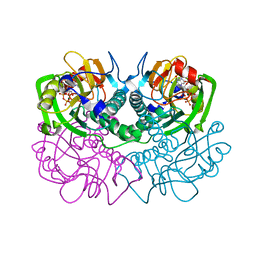 | | TOXOPLASMA GONDII HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE COMPLEXED WITH 9-DEAZAGUANINE, ALPHA-D-5-PHOSPHORIBOSYL-1-PYROPHOSPHATE (PRPP) AND TWO MG2+ IONS | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 9-DEAZAGUANINE, HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, ... | | Authors: | Heroux, A, White, E.L, Ross, L.J, Kuzin, A.P, Borhani, D.W. | | Deposit date: | 2000-09-08 | | Release date: | 2000-12-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Substrate deformation in a hypoxanthine-guanine phosphoribosyltransferase ternary complex: the structural basis for catalysis.
Structure Fold.Des., 8, 2000
|
|
1GCE
 
 | | STRUCTURE OF THE BETA-LACTAMASE OF ENTEROBACTER CLOACAE GC1 | | Descriptor: | BETA-LACTAMASE | | Authors: | Crichlow, G.V, Kuzin, A.P, Nukaga, M, Sawai, T, Knox, J.R. | | Deposit date: | 1999-05-17 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the extended-spectrum class C beta-lactamase of Enterobacter cloacae GC1, a natural mutant with a tandem tripeptide insertion.
Biochemistry, 38, 1999
|
|
2AXP
 
 | | X-Ray Crystal Structure of Protein BSU20280 from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR256. | | Descriptor: | hypothetical protein BSU20280 | | Authors: | Forouhar, F, Abashidze, M, Friedman, D, Vorobiev, S.M, Kuzin, A.P, Ciao, M, Conover, K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-09-05 | | Release date: | 2005-09-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Hypothetical Protein BSU20280 from Bacillus subtilis, NESG target SR256
To be Published
|
|
3NZL
 
 | | Crystal Structure of the N-terminal domain of DNA-binding protein SATB1 from Homo sapiens, Northeast Structural Genomics Consortium Target HR4435B | | Descriptor: | DNA-binding protein SATB1 | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Kuzin, A.P, Patel, P, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-07-16 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Crystal Structure of the N-terminal domain of DNA-binding protein SATB1 from Homo sapiens, Northeast Structural Genomics Consortium Target HR4435B (CASP Target)
To be Published
|
|
1NPD
 
 | | X-RAY STRUCTURE OF SHIKIMATE DEHYDROGENASE COMPLEXED WITH NAD+ FROM E.COLI (YDIB) NORTHEAST STRUCTURAL GENOMICS RESEARCH CONSORTIUM (NESG) TARGET ER24 | | Descriptor: | HYPOTHETICAL SHIKIMATE 5-DEHYDROGENASE-LIKE PROTEIN YDIB, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Benach, J, Kuzin, A.P, Lee, I, Rost, B, Chiang, Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-01-17 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 2.3-A crystal structure of the shikimate 5-dehydrogenase orthologue YdiB from Escherichia coli suggests a novel catalytic environment for an NAD-dependent dehydrogenase
J.Biol.Chem., 278, 2003
|
|
3PTE
 
 | |
3CFU
 
 | | Crystal structure of the yjhA protein from Bacillus subtilis. Northeast Structural Genomics Consortium target SR562 | | Descriptor: | Uncharacterized lipoprotein yjhA | | Authors: | Vorobiev, S.M, Chen, Y, Kuzin, A.P, Seetharaman, J, Forouhar, F, Zhao, L, Mao, L, Maglaqui, M, Xiao, R, Liu, J, Swapna, G, Huang, J.Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the yjhA protein from Bacillus subtilis.
To be Published
|
|
3D0W
 
 | | Crystal structure of YflH protein from Bacillus subtilis. Northeast Structural Genomics Consortium target SR326 | | Descriptor: | YflH protein | | Authors: | Seetharaman, J, Kuzin, A.P, Neely, H, Forouhar, F, Min, S, Zhao, L, Fang, Y, Owens, L, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-05-02 | | Release date: | 2008-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of YflH protein from Bacillus subtilis.
To be Published
|
|
1U1W
 
 | | Structure and function of phenazine-biosynthesis protein PhzF from Pseudomonas fluorescens 2-79 | | Descriptor: | 3-HYDROXYANTHRANILIC ACID, ACETATE ION, GLYCEROL, ... | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1U1X
 
 | | Structure and function of phenazine-biosynthesis protein PhzF from Pseudomonas fluorescens 2-79 | | Descriptor: | (2S,3S)-TRANS-2,3-DIHYDRO-3-HYDROXYANTHRANILIC ACID, Phenazine biosynthesis protein phzF | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1U1V
 
 | | Structure and function of phenazine-biosynthesis protein PhzF from Pseudomonas fluorescens 2-79 | | Descriptor: | GLYCEROL, Phenazine biosynthesis protein phzF, SULFATE ION | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2QTI
 
 | | Crystal structure of the UPF0352 protein SO_2176 from Shewanella oneidensis. NESG target SoR77. | | Descriptor: | UPF0352 protein SO_2176 | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Kuzin, A.P, Wang, D, Cunningham, K, Owens, L, Maglaqui, M, Fang, Y, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-08-02 | | Release date: | 2007-08-21 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the UPF0352 protein SO_2176 from Shewanella oneidensis.
To be Published
|
|
