6E9L
 
 | |
6SNJ
 
 | |
5FD6
 
 | |
8DYI
 
 | | IL17A homodimer bound to Compound 5 | | Descriptor: | (5P)-5-[5-(benzylamino)pyridin-3-yl]-N-[2-(morpholin-4-yl)ethyl]-1H-indazol-3-amine, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
8DYH
 
 | | IL17A homodimer bound to Compound 6 | | Descriptor: | (5P)-N-benzyl-6-chloro-5-(quinolin-5-yl)pyridin-3-amine, GLYCEROL, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
8DYF
 
 | | IL17A homodimer bound to Compound 10 | | Descriptor: | (5M)-3-[({2-[2-(2-{2-[2-({[(5M)-3-carboxy-5-(5,8-dihydroquinolin-4-yl)phenyl]amino}methyl)phenoxy]ethoxy}ethoxy)ethoxy]phenyl}methyl)amino]-5-(quinolin-4-yl)benzoic acid, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
9FGP
 
 | |
9GYL
 
 | | Ferredoxin Wild-type -Oxidised state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-1, chloroplastic, ... | | Authors: | Carr, S.B, Wei, J, Vincent, K.A. | | Deposit date: | 2024-10-02 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Cyanophenylalanine as an Infrared Probe for Iron-Sulfur Cluster Redox State in Multi-Centre Metalloenzymes.
Chembiochem, 2025
|
|
9GYN
 
 | | Ferredoxin Wild-type - Reduced state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-1, chloroplastic | | Authors: | Carr, S.B, Wei, J, Vincent, K.A. | | Deposit date: | 2024-10-02 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Cyanophenylalanine as an Infrared Probe for Iron-Sulfur Cluster Redox State in Multi-Centre Metalloenzymes.
Chembiochem, 2025
|
|
9GYR
 
 | | Ferredoxin CNF labelled, oxidised state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-1, chloroplastic | | Authors: | Carr, S.B, Wei, J, Vincent, K.A. | | Deposit date: | 2024-10-02 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Cyanophenylalanine as an Infrared Probe for Iron-Sulfur Cluster Redox State in Multi-Centre Metalloenzymes.
Chembiochem, 2025
|
|
9GYD
 
 | | Ferredoxin Wild-type -As-isolated state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-1, chloroplastic | | Authors: | Carr, S.B, Wei, J, Vincent, K.A. | | Deposit date: | 2024-10-01 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Cyanophenylalanine as an Infrared Probe for Iron-Sulfur Cluster Redox State in Multi-Centre Metalloenzymes.
Chembiochem, 2025
|
|
9GYU
 
 | | Ferredoxin CNF-labelled reduced state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-1, chloroplastic | | Authors: | Carr, S.B, Wei, J, Vincent, K.A. | | Deposit date: | 2024-10-03 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Cyanophenylalanine as an Infrared Probe for Iron-Sulfur Cluster Redox State in Multi-Centre Metalloenzymes.
Chembiochem, 2025
|
|
9GZ0
 
 | | FeFe Hydrogenase from Desulfovibrio desulfuricans labelled with cyanophenylalanine - oxidised state | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, LITHIUM ION, ... | | Authors: | Carr, S.B, Duan, Z, Rodroguez-Macia, P, Vincent, K.A. | | Deposit date: | 2024-10-03 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Cyanophenylalanine as an Infrared Probe for Iron-Sulfur Cluster Redox State in Multi-Centre Metalloenzymes.
Chembiochem, 2025
|
|
9GZ4
 
 | | FeFe Hydrogenase from Desulfovibrio desulfuricans labelled with cyanophenylalanine - reduced state | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, LITHIUM ION, ... | | Authors: | Carr, S.B, Duan, Z, Rodroguez-Macia, P, Vincent, K.A. | | Deposit date: | 2024-10-03 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Cyanophenylalanine as an Infrared Probe for Iron-Sulfur Cluster Redox State in Multi-Centre Metalloenzymes.
Chembiochem, 2025
|
|
9GZL
 
 | | Apo FeFe Hydrogenase from Desulfovibrio desulfuricans labelled with cyanophenylalanine | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, LITHIUM ION, ... | | Authors: | Carr, S.B, Duan, Z, Rodriguez-Macia, P, Vincent, K.A. | | Deposit date: | 2024-10-04 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Cyanophenylalanine as an Infrared Probe for Iron-Sulfur Cluster Redox State in Multi-Centre Metalloenzymes.
Chembiochem, 2025
|
|
4CNS
 
 | | Crystal structure of truncated human CRMP-4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ponnusamy, R, Lohkamp, B. | | Deposit date: | 2014-01-24 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Human Crmp-4: Correction of Intensities for Lattice-Translocation Disorder
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CNT
 
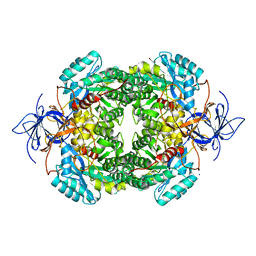 | | CRYSTAL STRUCTURE OF WT HUMAN CRMP-4 | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROPYRIMIDINASE-LIKE 3, SODIUM ION | | Authors: | Ponnusamy, R, Lohkamp, B. | | Deposit date: | 2014-01-24 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of Human Crmp-4: Correction of Intensities for Lattice-Translocation Disorder
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2GA2
 
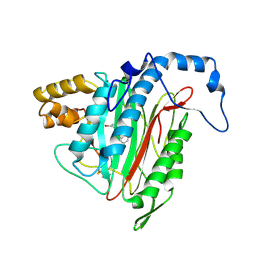 | | h-MetAP2 complexed with A193400 | | Descriptor: | 5-BROMO-2-{[(4-CHLOROPHENYL)SULFONYL]AMINO}BENZOIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C. | | Deposit date: | 2006-03-07 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development of sulfonamide compounds as potent methionine aminopeptidase type II inhibitors with antiproliferative properties.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3OHN
 
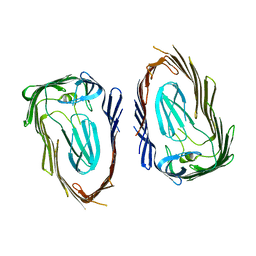 | | Crystal structure of the FimD translocation domain | | Descriptor: | Outer membrane usher protein FimD | | Authors: | Wang, T, Li, H. | | Deposit date: | 2010-08-17 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate.
Nature, 474, 2011
|
|
2M5Y
 
 | |
5UPE
 
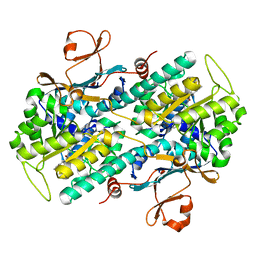 | |
5VHB
 
 | |
5VIB
 
 | |
5UPF
 
 | |
5VI9
 
 | |
