5HP8
 
 | | Crystal structures of RidA in complex with pyruvate | | Descriptor: | PYRUVIC ACID, Reactive Intermediate Deaminase A, chloroplastic | | Authors: | Xie, W, Liu, X. | | Deposit date: | 2016-01-20 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of RidA, an important enzyme for the prevention of toxic side products
Sci Rep, 6, 2016
|
|
5ZOV
 
 | | Inward-facing conformation of L-ascorbate transporter UlaA | | Descriptor: | ASCORBIC ACID, CALCIUM ION, PTS ascorbate-specific subunit IIBC | | Authors: | Wang, J.W. | | Deposit date: | 2018-04-16 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.333 Å) | | Cite: | Inward-facing conformation of l-ascorbate transporter suggests an elevator mechanism
Cell Discov, 4, 2018
|
|
5HP7
 
 | | Crystal structures of RidA in the apo form | | Descriptor: | Reactive Intermediate Deaminase A, chloroplastic | | Authors: | Xie, W, Liu, X. | | Deposit date: | 2016-01-20 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of RidA, an important enzyme for the prevention of toxic side products
Sci Rep, 6, 2016
|
|
6IER
 
 | | Apo structure of a beta-glucosidase 1317 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, beta-glucosidase 1317 | | Authors: | Xie, W, Liu, X. | | Deposit date: | 2018-09-16 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Improving the cellobiose-hydrolysis activity and glucose-tolerance of a thermostable beta-glucosidase through rational design.
Int.J.Biol.Macromol., 136, 2019
|
|
1CQE
 
 | | PROSTAGLANDIN H2 SYNTHASE-1 COMPLEX WITH FLURBIPROFEN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLURBIPROFEN, PROTEIN (PROSTAGLANDIN H2 SYNTHASE-1), ... | | Authors: | Picot, D, Loll, P.J, Mulichak, A.M, Garavito, R.M. | | Deposit date: | 1999-06-15 | | Release date: | 1999-06-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The X-ray crystal structure of the membrane protein prostaglandin H2 synthase-1.
Nature, 367, 1994
|
|
7WN6
 
 | | Cryo-EM structure of VWF D'D3 dimer (R1136M/E1143M mutant) complexed with D1D2 at 3.29 angstron resolution (2 units) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-17 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural mechanism of VWF D'D3 dimer formation.
Cell Discov, 8, 2022
|
|
7WN4
 
 | | Cryo-EM structure of VWF D'D3 dimer (wild type) complexed with D1D2 at 3.4 angstron resolution (1 unit) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-17 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanism of VWF D'D3 dimer formation.
Cell Discov, 8, 2022
|
|
7WN3
 
 | | Cryo-EM structure of VWF D'D3 dimer (2M mutant) complexed with D1D2 at 3.29 angstron resolution (2 units) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-17 | | Release date: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural mechanism of VWF D'D3 dimer formation.
Cell Discov, 8, 2022
|
|
7D0F
 
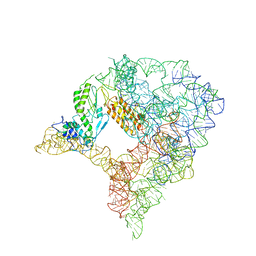 | | cryo-EM structure of a pre-catalytic group II intron RNP | | Descriptor: | Group II intron-encoded protein LtrA, RNA (738-MER) | | Authors: | Liu, N, Dong, X.L, Hu, C.X, Zeng, J.W, Wang, J.W, Wang, J, Wang, H.W, Belfort, M. | | Deposit date: | 2020-09-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Exon and protein positioning in a pre-catalytic group II intron RNP primed for splicing.
Nucleic Acids Res., 48, 2020
|
|
7D0G
 
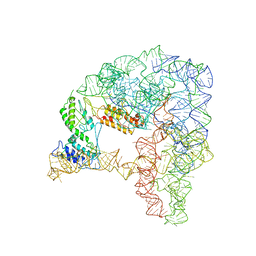 | | Cryo-EM structure of a pre-catalytic group II intron | | Descriptor: | Group II intron-encoded protein LtrA, RNA (714-MER) | | Authors: | Liu, N, Dong, X.L, Hu, C.X, Zeng, J.W, Wang, J.W, Wang, J, Wang, H.W, Belfort, M. | | Deposit date: | 2020-09-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Exon and protein positioning in a pre-catalytic group II intron RNP primed for splicing.
Nucleic Acids Res., 48, 2020
|
|
7D1A
 
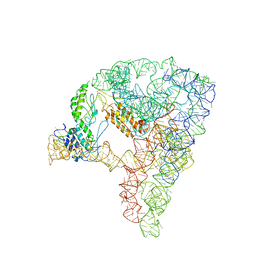 | | cryo-EM structure of a group II intron RNP complexed with its reverse transcriptase | | Descriptor: | Group II intron-encoded protein LtrA, RNA (5'-R(P*CP*AP*CP*AP*UP*CP*CP*AP*UP*AP*AP*C)-3'), RNA (692-MER) | | Authors: | Liu, N, Dong, X.L, Hu, C.X, Zeng, J.W, Wang, J.W, Wang, J, Wang, H.W, Belfort, M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Exon and protein positioning in a pre-catalytic group II intron RNP primed for splicing.
Nucleic Acids Res., 48, 2020
|
|
7DYR
 
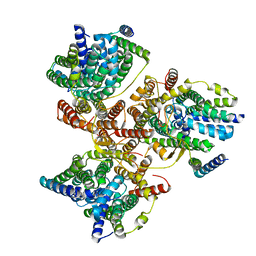 | |
8GU7
 
 | |
8GT9
 
 | |
7VEH
 
 | | Type I-F Anti-CRISPR protein AcrIF13 | | Descriptor: | AcrIF13 | | Authors: | Gao, T, Feng, Y. | | Deposit date: | 2021-09-08 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Mechanistic insights into the inhibition of the CRISPR-Cas surveillance complex by anti-CRISPR protein AcrIF13.
J.Biol.Chem., 298, 2022
|
|
5WT1
 
 | |
7VLX
 
 | | Cryo-EM structures of Listeria monocytogenes man-PTS | | Descriptor: | Mannose/fructose/sorbose family PTS transporter subunit IIC, PTS mannose family transporter subunit IID, alpha-D-mannopyranose | | Authors: | Wang, J.W. | | Deposit date: | 2021-10-05 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural Basis of Pore Formation in the Mannose Phosphotransferase System by Pediocin PA-1.
Appl.Environ.Microbiol., 88, 2022
|
|
7VLY
 
 | | Cryo-EM structure of Listeria monocytogenes man-PTS complexed with pediocin PA-1 | | Descriptor: | Bacteriocin pediocin PA-1, Mannose/fructose/sorbose family PTS transporter subunit IIC, PTS mannose family transporter subunit IID, ... | | Authors: | Wang, J.W. | | Deposit date: | 2021-10-05 | | Release date: | 2021-12-01 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural Basis of Pore Formation in the Mannose Phosphotransferase System by Pediocin PA-1.
Appl.Environ.Microbiol., 88, 2022
|
|
7VI9
 
 | |
7VII
 
 | |
7VIA
 
 | |
7VIK
 
 | |
7XNO
 
 | |
7WE6
 
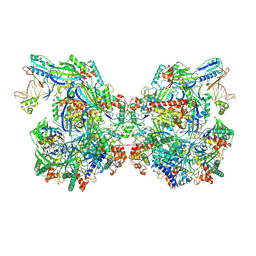 | | Structure of Csy-AcrIF24-dsDNA | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-12-22 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
7FI4
 
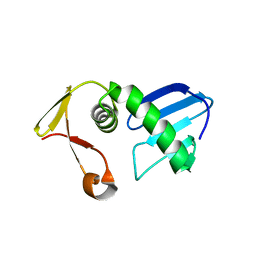 | | Structure of AcrIF13 | | Descriptor: | AcrIF13 | | Authors: | Feng, Y, Gao, T. | | Deposit date: | 2021-07-30 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Mechanistic insights into the inhibition of the CRISPR-Cas surveillance complex by anti-CRISPR protein AcrIF13.
J.Biol.Chem., 298, 2022
|
|
