2Y28
 
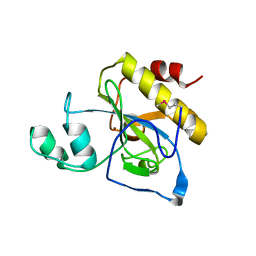 | | crystal structure of Se-Met AmpD derivative | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y2B
 
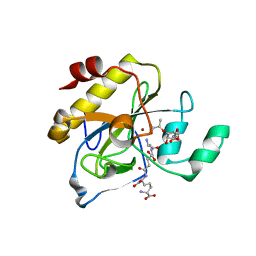 | | crystal structure of AmpD in complex with reaction products | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, L-ALA-GAMMA-D-GLU-MESO-DIAMINOPIMELIC ACID, ... | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
3ITA
 
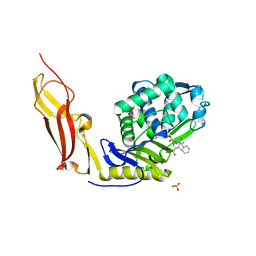 | | Crystal structure of Penicillin-Binding Protein 6 (PBP6) from E. coli in acyl-enzyme complex with ampicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, D-alanyl-D-alanine carboxypeptidase dacC, ... | | Authors: | Chen, Y, Zhang, W, Shi, Q, Hesek, D, Lee, M, Mobashery, S, Shoichet, B.K. | | Deposit date: | 2009-08-27 | | Release date: | 2009-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of penicillin-binding protein 6 from Escherichia coli.
J.Am.Chem.Soc., 131, 2009
|
|
2Y2E
 
 | | crystal structure of AmpD grown at pH 5.5 | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
3IT9
 
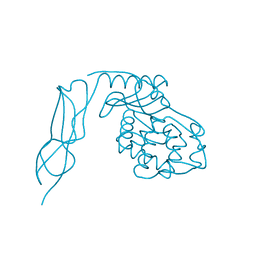 | | Crystal structure of Penicillin-Binding Protein 6 (PBP6) from E. coli in apo state | | Descriptor: | D-alanyl-D-alanine carboxypeptidase dacC, SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Chen, Y, Zhang, W, Shi, Q, Hesek, D, Lee, M, Mobashery, S, Shoichet, B.K. | | Deposit date: | 2009-08-27 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of penicillin-binding protein 6 from Escherichia coli.
J.Am.Chem.Soc., 131, 2009
|
|
3ITB
 
 | | Crystal structure of Penicillin-Binding Protein 6 (PBP6) from E. coli in complex with a substrate fragment | | Descriptor: | D-alanyl-D-alanine carboxypeptidase DacC, Peptidoglycan substrate (AMV)A(FGA)K(DAL)(DAL), SULFATE ION, ... | | Authors: | Chen, Y, Zhang, W, Shi, Q, Hesek, D, Lee, M, Mobashery, S, Shoichet, B.K. | | Deposit date: | 2009-08-27 | | Release date: | 2009-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of penicillin-binding protein 6 from Escherichia coli.
J.Am.Chem.Soc., 131, 2009
|
|
2Y2D
 
 | | crystal structure of AmpD holoenzyme | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
4MHA
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC1817 | | Descriptor: | 2-(butylamino)-4-[(trans-4-hydroxycyclohexyl)amino]-N-(4-sulfamoylbenzyl)pyrimidine-5-carboxamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Zhang, W, Mciver, A, Stashko, M.A, Deryckere, D, Branchford, B.R, Hunter, D, Kireev, D.B, Miley, D.B.M, Norris-Drouin, J, Stewart, W.M, Lee, M, Sather, S, Zhou, Y, Dipaola, J.A, Machius, M, Janzen, W.P, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2013-08-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Discovery of Mer specific tyrosine kinase inhibitors for the treatment and prevention of thrombosis.
J.Med.Chem., 56, 2013
|
|
6LCI
 
 | | Solution structure of mdaA-1 domain | | Descriptor: | mdaA-1 | | Authors: | Nguyen, T.A, Le, S, Lee, M, Fan, J.S, Yang, D, Yan, J, Jedd, G. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Fungal Wound Healing through Instantaneous Protoplasmic Gelation.
Curr.Biol., 31, 2021
|
|
7SMR
 
 | | Cryo-EM structure of Torpedo acetylcholine receptor in complex with carbachol, desensitized state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[(AMINOCARBONYL)OXY]-N,N,N-TRIMETHYLETHANAMINIUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rahman, M.M, Basta, T, Teng, J, Lee, M, Worrell, B.T, Stowell, M.H.B, Hibbs, R.E. | | Deposit date: | 2021-10-26 | | Release date: | 2022-03-09 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural mechanism of muscle nicotinic receptor desensitization and block by curare.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SMM
 
 | | Cryo-EM structure of Torpedo acetylcholine receptor in apo form | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rahman, M.M, Basta, T, Teng, J, Lee, M, Worrell, B.T, Stowell, M.H.B, Hibbs, R.E. | | Deposit date: | 2021-10-26 | | Release date: | 2022-03-09 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural mechanism of muscle nicotinic receptor desensitization and block by curare.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SMQ
 
 | | Cryo-EM structure of Torpedo acetylcholine receptor in apo form with added cholesterol | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rahman, M.M, Basta, T, Teng, J, Lee, M, Worrell, B.T, Stowell, M.H.B, Hibbs, R.E. | | Deposit date: | 2021-10-26 | | Release date: | 2022-03-09 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mechanism of muscle nicotinic receptor desensitization and block by curare.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SMS
 
 | | Cryo-EM structure of Torpedo acetylcholine receptor in complex with d-tubocurarine | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine receptor subunit alpha, ... | | Authors: | Rahman, M.M, Basta, T, Teng, J, Lee, M, Worrell, B.T, Stowell, M.H.B, Hibbs, R.E. | | Deposit date: | 2021-10-26 | | Release date: | 2022-03-09 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural mechanism of muscle nicotinic receptor desensitization and block by curare.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SMT
 
 | | Cryo-EM structure of Torpedo acetylcholine receptor in complex with d-tubocurarine and carbachol | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[(AMINOCARBONYL)OXY]-N,N,N-TRIMETHYLETHANAMINIUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rahman, M.M, Basta, T, Teng, J, Lee, M, Worrell, B.T, Stowell, M.H.B, Hibbs, R.E. | | Deposit date: | 2021-10-26 | | Release date: | 2022-03-09 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural mechanism of muscle nicotinic receptor desensitization and block by curare.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3EPO
 
 | | Crystal structure of Caulobacter crescentus ThiC complexed with HMP-P | | Descriptor: | (4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL DIHYDROGEN PHOSPHATE, Thiamine biosynthesis protein thiC | | Authors: | Li, S, Chatterjee, A, Zhang, Y, Grove, T.L, Lee, M, Krebs, C, Booker, S.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2008-09-29 | | Release date: | 2008-10-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily
Nat.Chem.Biol., 4, 2008
|
|
3EPM
 
 | | Crystal structure of Caulobacter crescentus ThiC | | Descriptor: | 4-AMINO-5-HYDROXYMETHYL-2-METHYLPYRIMIDINE, SULFATE ION, Thiamine biosynthesis protein thiC, ... | | Authors: | Li, S, Chatterjee, A, Zhang, Y, Grove, T.L, Lee, M, Krebs, C, Booker, S.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2008-09-29 | | Release date: | 2008-10-28 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily
Nat.Chem.Biol., 4, 2008
|
|
3EPN
 
 | | Crystal structure of Caulobacter crescentus ThiC complexed with imidazole ribonucleotide | | Descriptor: | 1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole, Thiamine biosynthesis protein thiC | | Authors: | Li, S, Chatterjee, A, Zhang, Y, Grove, T.L, Lee, M, Krebs, C, Booker, S.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2008-09-29 | | Release date: | 2008-10-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily
Nat.Chem.Biol., 4, 2008
|
|
3SEP
 
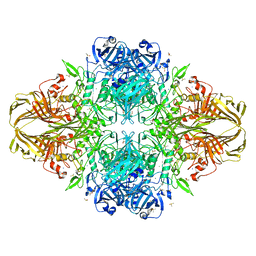 | | E. coli (lacZ) beta-galactosidase (S796A) | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-06-10 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3T0B
 
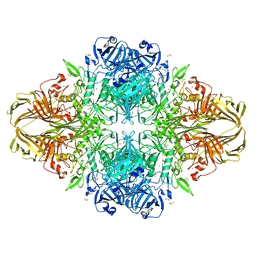 | | E. coli (LacZ) beta-galactosidase (S796T) IPTG complex | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-19 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | er-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
2MXB
 
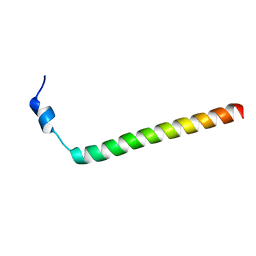 | |
3T2P
 
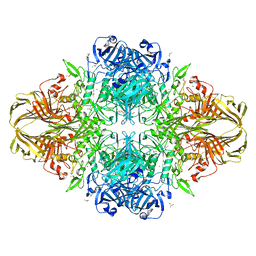 | | E. coli (lacZ) beta-galactosidase (S796D) in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-22 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
4UV4
 
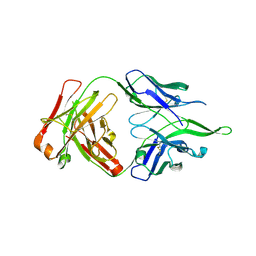 | | Crystal structure of anti-FPR Fpro0165 Fab fragment | | Descriptor: | FPRO0165 FAB | | Authors: | Douthwaite, J.A, Sridharan, S, Huntington, C, Marwood, R, Hammersley, J, Hakulinen, J.K, Ek, M, Sjogren, T, Rider, D, Privezentzev, C, Seaman, J.C, Cariuk, P, Knights, V, Young, J, Wilkinson, T, Sleeman, M, Finch, D.K, Lowe, D.C, Vaughan, T.J. | | Deposit date: | 2014-08-04 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Affinity Maturation of a Novel Antagonistic Human Monoclonal Antibody with a Long Vh Cdr3 Targeting the Class a Gpcr Formyl-Peptide Receptor 1.
Mabs, 7, 2015
|
|
3G9Y
 
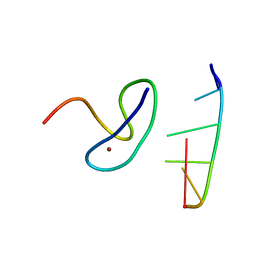 | | Crystal structure of the second zinc finger from ZRANB2/ZNF265 bound to 6 nt ssRNA sequence AGGUAA | | Descriptor: | RNA (5'-R(*AP*GP*GP*UP*AP*A)-3'), ZINC ION, Zinc finger Ran-binding domain-containing protein 2 | | Authors: | Loughlin, F.E, McGrath, A.P, Lee, M, Guss, J.M, Mackay, J.P. | | Deposit date: | 2009-02-15 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The zinc fingers of the SR-like protein ZRANB2 are single-stranded RNA-binding domains that recognize 5' splice site-like sequences
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1FUR
 
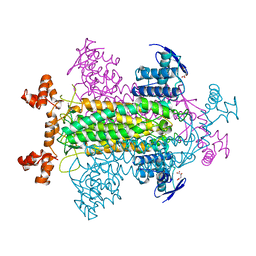 | |
2FUS
 
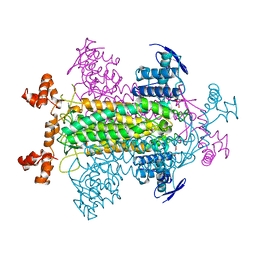 | |
