8HE1
 
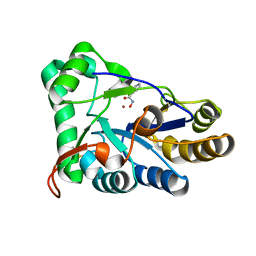 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | BENZHYDROXAMIC ACID, Chitin deacetylase, ZINC ION | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HFA
 
 | |
8HF9
 
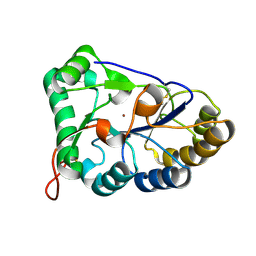 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-10 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
1QOB
 
 | | FERREDOXIN MUTATION D62K | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN, SULFATE ION | | Authors: | Holden, H.M, Benning, M.M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
1QOF
 
 | | FERREDOXIN MUTATION Q70K | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN, SULFATE ION | | Authors: | Holden, H.M, Benning, M.M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
1QOG
 
 | | FERREDOXIN MUTATION S47A | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN, SULFATE ION | | Authors: | Holden, H.M, Benning, M.M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
1QOA
 
 | | FERREDOXIN MUTATION C49S | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Holden, H.M, Benning, M.M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Iron-sulfur cluster cysteine-to-serine mutants of Anabaena -2Fe-2S- ferredoxin exhibit unexpected redox properties and are competent in electron transfer to ferredoxin:NADP+ reductase.
Biochemistry, 36, 1997
|
|
3MI8
 
 | | The structure of TL1A-DCR3 COMPLEX | | Descriptor: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 15, SECRETED FORM, Tumor necrosis factor receptor superfamily member 6B | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2010-04-09 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
3NUP
 
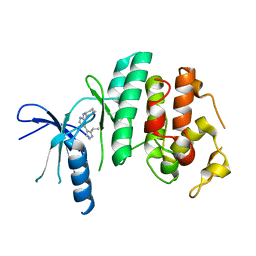 | | CDK6 (monomeric) in complex with inhibitor | | Descriptor: | 4-[3-(1-methylethyl)-1H-pyrazol-4-yl]-N-(1-methylpiperidin-4-yl)pyrimidin-2-amine, Cell division protein kinase 6 | | Authors: | Chopra, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 4-(Pyrazol-4-yl)-pyrimidines as selective inhibitors of cyclin-dependent kinase 4/6.
J.Med.Chem., 53, 2010
|
|
3NUX
 
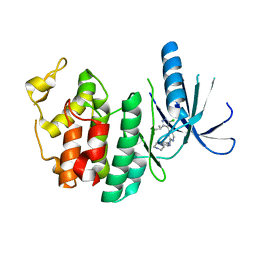 | | CDK6 (monomeric) in complex with inhibitor | | Descriptor: | 4-[5-chloro-3-(1-methylethyl)-1H-pyrazol-4-yl]-N-(5-piperazin-1-ylpyridin-2-yl)pyrimidin-2-amine, Cell division protein kinase 6 | | Authors: | Chopra, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 4-(Pyrazol-4-yl)-pyrimidines as selective inhibitors of cyclin-dependent kinase 4/6.
J.Med.Chem., 53, 2010
|
|
7V2Y
 
 | | cryo-EM structure of yeast THO complex with Sub2 | | Descriptor: | ATP-dependent RNA helicase SUB2, Protein TEX1, THO complex subunit 2, ... | | Authors: | Chen, C, Tan, M, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2021-08-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and functional insights into R-loop prevention and mRNA export by budding yeast THO-Sub2 complex.
Sci Bull (Beijing), 66, 2021
|
|
3FN3
 
 | | Dimeric Structure of PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Chen, Y, Gao, F, Liu, P, Chu, F, Qi, J, Gao, G.F. | | Deposit date: | 2008-12-23 | | Release date: | 2009-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A dimeric structure of PD-L1: functional units or evolutionary relics?
Protein Cell, 1, 2010
|
|
7V2W
 
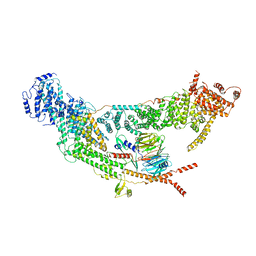 | | protomer structure from the dimer of yeast THO complex | | Descriptor: | Protein TEX1, THO complex subunit 2, THO complex subunit HPR1, ... | | Authors: | Chen, C, Tan, M, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2021-08-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional insights into R-loop prevention and mRNA export by budding yeast THO-Sub2 complex.
Sci Bull (Beijing), 66, 2021
|
|
6IHJ
 
 | |
7UV1
 
 | | Vicilin Ana o 1.0101 leader sequence residues 20-75 | | Descriptor: | Vicilin-like protein | | Authors: | Mueller, G.A, Foo, A.C.Y, DeRose, E.F. | | Deposit date: | 2022-04-29 | | Release date: | 2023-04-05 | | Method: | SOLUTION NMR | | Cite: | Structure and IgE Cross-Reactivity among Cashew, Pistachio, Walnut, and Peanut Vicilin-Buried Peptides.
J.Agric.Food Chem., 71, 2023
|
|
7UV4
 
 | | Pis v 3.0101 vicilin leader sequence residues 56-115 | | Descriptor: | Vicilin Pis v 3.0101 | | Authors: | Mueller, G.A, Foo, A.C.Y, DeRose, E.F. | | Deposit date: | 2022-04-29 | | Release date: | 2023-04-05 | | Method: | SOLUTION NMR | | Cite: | Structure and IgE Cross-Reactivity among Cashew, Pistachio, Walnut, and Peanut Vicilin-Buried Peptides.
J.Agric.Food Chem., 71, 2023
|
|
7UV2
 
 | | Ana o 1 Leader Sequence Residues 82-132 | | Descriptor: | Vicilin-like protein | | Authors: | Mueller, G.A, Foo, A.C.Y, DeRose, E.F. | | Deposit date: | 2022-04-29 | | Release date: | 2023-04-05 | | Method: | SOLUTION NMR | | Cite: | Structure and IgE Cross-Reactivity among Cashew, Pistachio, Walnut, and Peanut Vicilin-Buried Peptides.
J.Agric.Food Chem., 71, 2023
|
|
7UV3
 
 | | Pis v 3.0101 Vicilin Leader Sequence Residues 5-52 | | Descriptor: | Vicilin Pis v 3.0101 | | Authors: | Mueller, G.A, Foo, A.C.Y, DeRose, E.F. | | Deposit date: | 2022-04-29 | | Release date: | 2023-04-05 | | Method: | SOLUTION NMR | | Cite: | Structure and IgE Cross-Reactivity among Cashew, Pistachio, Walnut, and Peanut Vicilin-Buried Peptides.
J.Agric.Food Chem., 71, 2023
|
|
3PRE
 
 | | Quinazolines with intra-molecular hydrogen bonding scaffold (iMHBS) as PI3K/mTOR dual inhibitors. | | Descriptor: | 2-amino-8-(trans-4-methoxycyclohexyl)-4-methyl-6-(1H-pyrazol-3-yl)pyrido[2,3-d]pyrimidin-7(8H)-one, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Knighton, D.R, Greasley, S.E, Rodgers, C.M.-L. | | Deposit date: | 2010-11-29 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Quinazolines with intra-molecular hydrogen bonding scaffold (iMHBS) as PI3K/mTOR dual inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3PRZ
 
 | |
3PS6
 
 | |
6VMZ
 
 | | Crystal Structure of a H5N1 influenza virus hemagglutinin with CBS1117 | | Descriptor: | 2,6-dichloro-N-[1-(propan-2-yl)piperidin-4-yl]benzamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Antanasijevic, A, Durst, M.A, Lavie, A, Caffrey, M. | | Deposit date: | 2020-01-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of avian influenza hemagglutinin in complex with a small molecule entry inhibitor.
Life Sci Alliance, 3, 2020
|
|
4EN0
 
 | | Crystal structure of light | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Zhan, C, Liu, W, Patskovsky, Y, Ramagopal, U.A, Bonanno, J.B, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
4J6G
 
 | | CRYSTAL STRUCTURE OF LIGHT AND DcR3 COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Liu, W, Zhan, C, Bonanno, J.B, Bhosle, R.C, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-11 | | Release date: | 2013-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
3UDD
 
 | | Tankyrase-1 in complex with small molecule inhibitor | | Descriptor: | 3-(4-methoxyphenyl)-5-({[4-(4-methoxyphenyl)-5-methyl-4H-1,2,4-triazol-3-yl]sulfanyl}methyl)-1,2,4-oxadiazole, GLYCEROL, SULFATE ION, ... | | Authors: | Kirby, C.A, Stams, T. | | Deposit date: | 2011-10-28 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | [1,2,4]triazol-3-ylsulfanylmethyl)-3-phenyl-[1,2,4]oxadiazoles: antagonists of the Wnt pathway that inhibit tankyrases 1 and 2 via novel adenosine pocket binding.
J.Med.Chem., 55, 2012
|
|
