7EYP
 
 | | Crystal structure of Pseudomonas aeruginosa ppnP | | Descriptor: | Pyrimidine/purine nucleoside phosphorylase | | Authors: | Wen, Y, Wu, B.X. | | Deposit date: | 2021-05-31 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of a new class of pyrimidine/purine nucleoside phosphorylase revealed a Cupin fold.
Proteins, 90, 2022
|
|
7EYJ
 
 | | Crystal structure of Escherichia coli ppnP | | Descriptor: | Pyrimidine/purine nucleoside phosphorylase, SULFATE ION | | Authors: | Wen, Y, Wu, B.X. | | Deposit date: | 2021-05-31 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structures of a new class of pyrimidine/purine nucleoside phosphorylase revealed a Cupin fold.
Proteins, 90, 2022
|
|
7EYL
 
 | | Crystal structure of Salmonella enterica ppnP | | Descriptor: | Pyrimidine/purine nucleoside phosphorylase | | Authors: | Wen, Y, Wu, B.X. | | Deposit date: | 2021-05-31 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of a new class of pyrimidine/purine nucleoside phosphorylase revealed a Cupin fold.
Proteins, 90, 2022
|
|
7EYM
 
 | | Crystal structure of Vibrio cholerae ppnP | | Descriptor: | Pyrimidine/purine nucleoside phosphorylase | | Authors: | Wen, Y, Wu, B.X. | | Deposit date: | 2021-05-31 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structures of a new class of pyrimidine/purine nucleoside phosphorylase revealed a Cupin fold.
Proteins, 90, 2022
|
|
8J0D
 
 | | FCP heterodimer, Lhca2, and Lhcf5 together as the M1 side binds to the PSII core in the diatom Thalassiosira pseudonana | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Li, Z, Feng, Y, Wang, W, Shen, J.R. | | Deposit date: | 2023-04-10 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structure of a diatom photosystem II supercomplex containing a member of Lhcx family and dimeric FCPII.
Sci Adv, 9, 2023
|
|
7EQT
 
 | |
5K23
 
 | |
7EQS
 
 | |
7EQW
 
 | |
7ER1
 
 | |
7ER0
 
 | |
3E17
 
 | | Crystal structure of the second PDZ domain from human Zona Occludens-2 | | Descriptor: | Tight junction protein ZO-2 | | Authors: | Chen, H, Tong, S.L, Teng, M.K, Niu, L.W. | | Deposit date: | 2008-08-01 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the second PDZ domain from human zonula occludens 2
Acta Crystallogr.,Sect.F, 65, 2009
|
|
8XKN
 
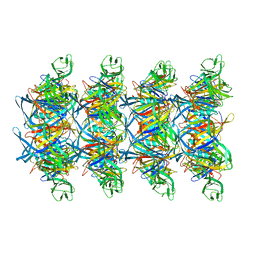 | | Cryo-EM structure of tail tube protein | | Descriptor: | a protein | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-12-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
4B4L
 
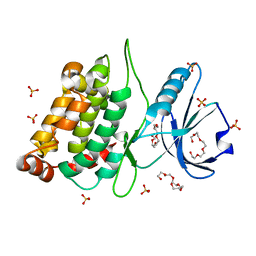 | | CRYSTAL STRUCTURE OF AN ARD DAP-KINASE 1 MUTANT | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE 1, PENTAETHYLENE GLYCOL, SULFATE ION | | Authors: | Temmerman, K, Pogenberg, V, Jonko, W, Wilmanns, M. | | Deposit date: | 2012-07-31 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Pef/Y Substrate Recognition and Signature Motif Plays a Critical Role in Dapk-Related Kinase Activity.
Chem.Biol., 21, 2014
|
|
5MOY
 
 | |
5NDF
 
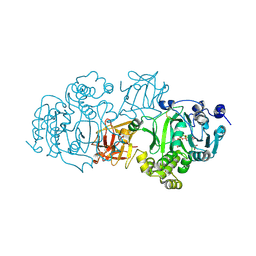 | | Small-molecule inhibition of ppGalNAc-Ts selectively reduces mucin-type O-glycosylation | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, MANGANESE (II) ION, ... | | Authors: | Hurtado-Guerrero, R, De las Rivas, M. | | Deposit date: | 2017-03-08 | | Release date: | 2017-11-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The small molecule luteolin inhibits N-acetyl-alpha-galactosaminyltransferases and reduces mucin-type O-glycosylation of amyloid precursor protein.
J. Biol. Chem., 292, 2017
|
|
3EZX
 
 | |
5V7U
 
 | |
6KJY
 
 | | Galectin-13 variant C136S/C138S | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Su, J. | | Deposit date: | 2019-07-23 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Galectin-13/placental protein 13: redox-active disulfides as switches for regulating structure, function and cellular distribution.
Glycobiology, 30, 2020
|
|
3ZDP
 
 | |
6KJW
 
 | | Galectin-13 variant C136S | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Su, J. | | Deposit date: | 2019-07-23 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Galectin-13/placental protein 13: redox-active disulfides as switches for regulating structure, function and cellular distribution.
Glycobiology, 30, 2020
|
|
4KFW
 
 | |
5V7R
 
 | |
3FW5
 
 | |
3FW4
 
 | |
