8R55
 
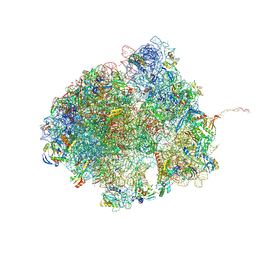 | | Bacillus subtilis MutS2-collided disome complex (collided 70S) | | Descriptor: | 16S rRNA (1533-MER), 23S RNA (2887-MER), 30S ribosomal protein S10, ... | | Authors: | Park, E, Mackens-Kiani, T, Berhane, R, Esser, H, Erdenebat, C, Burroughs, A.M, Berninghausen, O, Aravind, L, Beckmann, R, Green, R, Buskirk, A.R. | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | B. subtilis MutS2 splits stalled ribosomes into subunits without mRNA cleavage.
Embo J., 43, 2024
|
|
4BUI
 
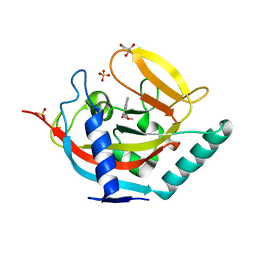 | | Crystal structure of human tankyrase 2 in complex with methyl 4-(4- oxo-3,4-dihydroquinazolin-2-yl)benzoate | | Descriptor: | GLYCEROL, METHYL 4-(4-OXO-3,4-DIHYDROQUINAZOLIN-2-YL)BENZOATE, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Para-Substituted 2-Phenyl-3,4-Dihydroquinazolin-4-Ones as Potent and Selective Tankyrase Inhibitors.
Chemmedchem, 8, 2013
|
|
3TDM
 
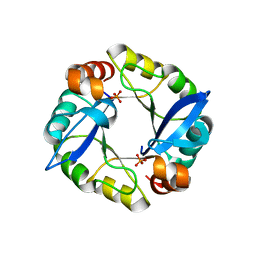 | | Computationally designed TIM-barrel protein, HalfFLR | | Descriptor: | Computationally designed two-fold symmetric TIM-barrel protein, FLR (half molecule), PHOSPHATE ION | | Authors: | Harp, J.M, Fortenberry, C, Bowman, E, Profitt, W, Dorr, B, Mizoue, L. | | Deposit date: | 2011-08-11 | | Release date: | 2011-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Exploring symmetry as an avenue to the computational design of large protein domains.
J.Am.Chem.Soc., 133, 2011
|
|
8UDV
 
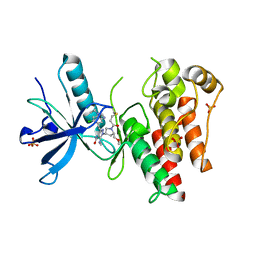 | | The X-RAY co-crystal structure of human FGFR3 V555M and Compound 17 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(6-chloro-1-cyclopropyl-1H-benzimidazol-5-yl)ethynyl]-1-[(3S,5S)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, Fibroblast growth factor receptor 3, ... | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
5MDT
 
 | |
4BUX
 
 | | Crystal structure of human tankyrase 2 in complex with 3-((4-(4-oxo-3, 4-dihydroquinazolin-2-yl)phenyl)methyl)imidazolidine-2,4-dione | | Descriptor: | 3-[[4-(4-oxidanylidene-3H-quinazolin-2-yl)phenyl]methyl]imidazolidine-2,4-dione, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-06-24 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Para-Substituted 2-Phenyl-3,4-Dihydroquinazolin-4-Ones as Potent and Selective Tankyrase Inhibitors.
Chemmedchem, 8, 2013
|
|
3TEB
 
 | | endonuclease/exonuclease/phosphatase family protein from Leptotrichia buccalis C-1013-b | | Descriptor: | Endonuclease/exonuclease/phosphatase, MAGNESIUM ION | | Authors: | Chang, C, Bigelow, L, Muniez, I, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-08-12 | | Release date: | 2011-08-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structure of endonuclease/exonuclease/phosphatase family protein from Leptotrichia buccalis C-1013-b
To be Published
|
|
5YH5
 
 | | The crystal structure of Staphylococcus aureus CntA in apo form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Nickel ABC transporter substrate-binding protein | | Authors: | Song, L, Ji, Q. | | Deposit date: | 2017-09-27 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanistic insights into staphylopine-mediated metal acquisition
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8BV0
 
 | | Binary complex between the NB-ARC domain from the Tomato immune receptor NRC1 and the SPRY domain-containing effector SS15 from the potato cyst nematode | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NRC1, Truncated secreted SPRY domain-containing protein 15 (Fragment) | | Authors: | Contreras, M.P, Pai, H, Muniyandi, S, Toghani, A, Lawson, D.M, Tumtas, Y, Duggan, C, Yuen, E.L.H, Stevenson, C.E.M, Harant, A, Wu, C.H, Bozkurt, T.O, Kamoun, S, Derevnina, L. | | Deposit date: | 2022-12-01 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Resurrection of plant disease resistance proteins via helper NLR bioengineering.
Sci Adv, 9, 2023
|
|
4BP3
 
 | | Crystal Structure of Plasmodium Falciparum Spermidine Synthase in Complex with DECARBOXYLATED S-ADENOSYLMETHIONINE5' AND 4- METHYLANILINE | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 4-METHYLANILINE, 5'-[(S)-(3-AMINOPROPYL)(METHYL)-LAMBDA~4~-SULFANYL]-5'-DEOXYADENOSINE, ... | | Authors: | Sprenger, J, Halander, J.C, Svensson, B, Al-Karadaghi, S, Persson, L. | | Deposit date: | 2013-05-23 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Three-Dimensional Structures of Plasmodium Falciparum Spermidine Synthase with Bound Inhibitors Suggest New Strategies for Drug Design
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4C08
 
 | | Crystal structure of M. musculus protein arginine methyltransferase PRMT6 with CaCl2 at 1.34 Angstroms | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, PROTEIN ARGININE N-METHYLTRANSFERASE 6 | | Authors: | Bonnefond, L, Cura, V, Troffer-Charlier, N, Mailliot, J, Wurtz, J.M, Cavarelli, J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.338 Å) | | Cite: | Functional Insights from High Resolution Structures of Mouse Protein Arginine Methyltransferase 6.
J.Struct.Biol., 191, 2015
|
|
3TGJ
 
 | | S195A TRYPSINOGEN COMPLEXED WITH BOVINE PANCREATIC TRYPSIN INHIBITOR (BPTI) | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR, CALCIUM ION, SULFATE ION, ... | | Authors: | Pasternak, A, Ringe, D, Hedstrom, L. | | Deposit date: | 1998-07-16 | | Release date: | 1998-12-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparison of Anionic and Cationic Trypsinogens: The Anionic Activation Domain is More Flexible in Solution and Differs in its Mode of Bpti Binding in the Crystal Structure
Protein Sci., 8, 1999
|
|
8V1F
 
 | | TMPRSS2 complexed with the noncovalent inhibitor 6-amidino-2-napthol | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fraser, B.J, Dong, A, Kutera, M, Seitova, A, Li, Y, Hutchinson, A, Edwards, A, Benard, F, Halabelian, L, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-20 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | TMPRSS2 complexed with the noncovalent inhibitor 6-amidino-2-napthol
To Be Published
|
|
4BU8
 
 | | Crystal structure of human tankyrase 2 in complex with 4-(4-oxo-1,4- dihydroquinazolin-2-yl)benzonitrile | | Descriptor: | 4-(4-OXO-1,4-DIHYDROQUINAZOLIN-2-YL)BENZONITRILE, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Para-Substituted 2-Phenyl-3,4-Dihydroquinazolin-4-Ones as Potent and Selective Tankyrase Inhibitors.
Chemmedchem, 8, 2013
|
|
3T6F
 
 | | Biotin complex of Y54F core streptavidin | | Descriptor: | BIOTIN, BIOTIN-D-SULFOXIDE, GLYCEROL, ... | | Authors: | Baugh, L, Le Trong, I, Cerutti, D.S, Mehta, N, Gulich, S, Stayton, P.S, Stenkamp, R.E, Lybrand, T.P. | | Deposit date: | 2011-07-28 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Second-Contact Shell Mutation Diminishes Streptavidin-Biotin Binding Affinity through Transmitted Effects on Equilibrium Dynamics.
Biochemistry, 51, 2012
|
|
3T6L
 
 | | Y54F mutant of core streptavidin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Streptavidin | | Authors: | Baugh, L, Le Trong, I, Stayton, P.S, Stenkamp, R.E, Lybrand, T.P. | | Deposit date: | 2011-07-28 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Second-Contact Shell Mutation Diminishes Streptavidin-Biotin Binding Affinity through Transmitted Effects on Equilibrium Dynamics.
Biochemistry, 51, 2012
|
|
8UUK
 
 | | Pendrin in apo | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Wang, L, Hoang, A, Zhou, M. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of anion exchange and small-molecule inhibition of pendrin.
Nat Commun, 15, 2024
|
|
5MJ5
 
 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetichonokiol derivative 3 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[4-oxidanyl-3-[3-(phenylmethyl)phenyl]phenyl]prop-2-enoic acid, LYS-HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN-ASP-SER, Retinoic acid receptor RXR-alpha | | Authors: | Andrei, S.A, Brunsveld, L, Scheepstra, M, Ottmann, C. | | Deposit date: | 2016-11-30 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
1E7Z
 
 | | Crystal structure of the EMAP2/RNA binding domain of the p43 protein from human aminoacyl-tRNA synthetase complex | | Descriptor: | ENDOTHELIAL-MONOCYTE ACTIVATING POLYPEPTIDE II, MERCURY (II) ION | | Authors: | Pasqualato, S, Kerjan, P, Renault, L, Menetrey, J, Mirande, M, Cherfils, J. | | Deposit date: | 2000-09-13 | | Release date: | 2000-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the Emapii Domain of Human Aminoacyl-tRNA Synthetase Complex Reveals Evolutionary Dimeric Mimicry
Embo J., 20, 2001
|
|
5MQ1
 
 | | Crystal structure of the BRD7 bromodomain in complex with BI-9564 | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-[(dimethylamino)methyl]-2,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, Bromodomain-containing protein 7, ... | | Authors: | Diaz-Saez, L, Martin, L.J, Panagakou, I, Picaud, S, Krojer, T, von Delft, F, Knapp, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-12-19 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the BRD7 bromodomain in complex with BI-9564
To Be Published
|
|
5MQI
 
 | |
5YN6
 
 | | Crystal structure of MERS-CoV nsp10/nsp16 complex bound to SAM | | Descriptor: | S-ADENOSYLMETHIONINE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
4C8D
 
 | | Crystal structure of JmjC domain of human histone 3 Lysine-specific demethylase 3B (KDM3B) | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vollmar, M, Johansson, C, Gileadi, C, Goubin, S, Szykowska, A, Krojer, T, Crawley, L, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2013-09-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of Jmjc Domain of Human Histone 3 Lysine-Specific Demethylase 3B (Kdm3B)
To be Published
|
|
5YNN
 
 | | Crystal structure of MERS-CoV nsp16/nsp10complex bound to sinefungin and m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, SINEFUNGIN, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
6F27
 
 | | NMR solution structure of non-bound [des-Arg10]-kallidin (DAKD) | | Descriptor: | DAKD | | Authors: | Richter, C, Jonker, H.R.A, Schwalbe, H, Joedicke, L, Mao, J, Kuenze, G, Reinhart, C, Kalavacherla, T, Meiler, J, Preu, J, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-23 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
