8K4Q
 
 | |
8UUL
 
 | | Prototypic SARS-CoV-2 spike (containing K417) in the closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Geng, Q, Liu, B, Li, F. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Lys417 acts as a molecular switch that regulates the conformation of SARS-CoV-2 spike protein.
Elife, 12, 2023
|
|
8UUM
 
 | | Prototypic SARS-CoV-2 spike (containing K417) in the open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose, ... | | Authors: | Geng, Q, Liu, B, Li, F. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Lys417 acts as a molecular switch that regulates the conformation of SARS-CoV-2 spike protein.
Elife, 12, 2023
|
|
8UUN
 
 | | Prototypic SARS-CoV-2 spike (containing V417) in the closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Geng, Q, Liu, B, Li, F. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Lys417 acts as a molecular switch that regulates the conformation of SARS-CoV-2 spike protein.
Elife, 12, 2023
|
|
8UUO
 
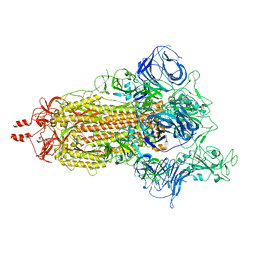 | | Prototypic SARS-CoV-2 spike (containing V417) in the open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose, ... | | Authors: | Geng, Q, Liu, B, Li, F. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Lys417 acts as a molecular switch that regulates the conformation of SARS-CoV-2 spike protein.
Elife, 12, 2023
|
|
4GH6
 
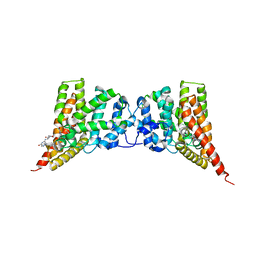 | | Crystal structure of the PDE9A catalytic domain in complex with inhibitor 28 | | Descriptor: | High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, N-(4-methoxyphenyl)-N~2~-[1-(2-methylphenyl)-4-oxo-4,5-dihydro-1H-pyrazolo[3,4-d]pyrimidin-6-yl]-L-alaninamide, ... | | Authors: | Hou, J, Ke, H. | | Deposit date: | 2012-08-07 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Based Discovery of Highly Selective Phosphodiesterase-9A Inhibitors and Implications for Inhibitor Design.
J.Med.Chem., 55, 2012
|
|
4QGE
 
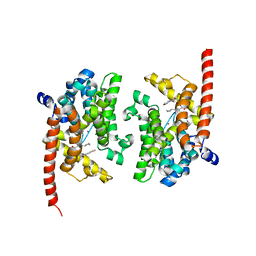 | | phosphodiesterase-9A in complex with inhibitor WYQ-C36D | | Descriptor: | MAGNESIUM ION, N~2~-(1-cyclopentyl-4-oxo-4,7-dihydro-1H-pyrazolo[3,4-d]pyrimidin-6-yl)-N-(4-methoxyphenyl)-D-alaninamide, Phosphodiesterase 9A, ... | | Authors: | Shao, Y.-X, Huang, M, Cui, W, Ke, H. | | Deposit date: | 2014-05-22 | | Release date: | 2014-12-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a Phosphodiesterase 9A Inhibitor as a Potential Hypoglycemic Agent.
J.Med.Chem., 57, 2014
|
|
7MBO
 
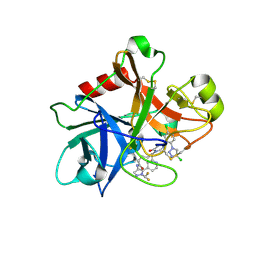 | | FACTOR XIA (PICHIA PASTORIS; C500S [C122S]) IN COMPLEX WITH THE INHIBITOR Milvexian (BMS-986177), IUPAC NAME:(6R,10S)-10-{4-[5-chloro-2-(4-chloro-1H-1,2,3-triazol-1-yl)phenyl]-6- oxopyrimidin-1(6H)-yl}-1-(difluoromethyl)-6-methyl-1,4,7,8,9,10-hexahydro-15,11- (metheno)pyrazolo[4,3-b][1,7]diazacyclotetradecin-5(6H)-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIa light chain, Milvexian | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.924 Å) | | Cite: | Discovery of Milvexian, a High-Affinity, Orally Bioavailable Inhibitor of Factor XIa in Clinical Studies for Antithrombotic Therapy.
J.Med.Chem., 65, 2022
|
|
5EIU
 
 | | Mini TRIM5 B-box 2 dimer C2 crystal form | | Descriptor: | TRIM protein-E3 ligase Chimera, ZINC ION | | Authors: | Wagner, J.M, Doss, G, Pornillos, O. | | Deposit date: | 2015-10-30 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | Mechanism of B-box 2 domain-mediated higher-order assembly of the retroviral restriction factor TRIM5 alpha.
Elife, 5, 2016
|
|
5F7T
 
 | | TRIM5 B-box2 and coiled-coil chimera | | Descriptor: | Tripartite motif-containing protein 5,Serine--tRNA ligase,Tripartite motif-containing protein 5, ZINC ION | | Authors: | Wagner, J.M, Doss, G, Pornillos, O. | | Deposit date: | 2015-12-08 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Mechanism of B-box 2 domain-mediated higher-order assembly of the retroviral restriction factor TRIM5 alpha.
Elife, 5, 2016
|
|
5VA4
 
 | |
8J1H
 
 | | Agonist1 and Ruthenium Red bound state of mTRPV4 | | Descriptor: | 8-fluoranyl-3-[4-(4-fluoranylphenoxy)phenyl]-2-(4-methylpiperazin-1-yl)quinazolin-4-one, Transient receptor potential cation channel subfamily V member 4 | | Authors: | Zhen, W.X, Yang, F. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
8J1D
 
 | | Cryo-EM structure of apo state mTRPV4 | | Descriptor: | Transient receptor potential cation channel subfamily V member 4 | | Authors: | Zhen, W.X, Yang, F. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
8J1F
 
 | | GSK101 bound state of mTRPV4 | | Descriptor: | N-[(2S)-1-{4-[N-(2,4-dichlorobenzene-1-sulfonyl)-L-seryl]piperazin-1-yl}-4-methyl-1-oxopentan-2-yl]-1-benzothiophene-2-carboxamide, Transient receptor potential cation channel subfamily V member 4 | | Authors: | Zhen, W.X, Yang, F. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
8J1B
 
 | | GSK101 and Ruthenium Red bound state of mTRPV4 | | Descriptor: | N-[(2S)-1-{4-[N-(2,4-dichlorobenzene-1-sulfonyl)-L-seryl]piperazin-1-yl}-4-methyl-1-oxopentan-2-yl]-1-benzothiophene-2-carboxamide, Transient receptor potential cation channel subfamily V member 4, ruthenium(6+) azanide pentaamino(oxido)ruthenium (1/4/2) | | Authors: | Zhen, W.X, Yang, F. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
2AEW
 
 | |
8JON
 
 | | Structure of a synthetic circadian clock protein KaiC mutant of cyanobacteria Synechococcus elongatus PCC 7942 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock oscillator protein KaiC, ... | | Authors: | Jia, X, Zhang, Q, Li, S, Guo, J. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Adaptation of ancient cyanobacterial clock to the day length ~ 0.95 Ga ago
To Be Published
|
|
5IEA
 
 | | TRIM5 B-box2 and coiled-coil chimera | | Descriptor: | Tripartite motif-containing protein 5, Serine--tRNA ligase Chimera, ZINC ION | | Authors: | Wagner, J.M, Doss, G, Pornillos, O. | | Deposit date: | 2016-02-25 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.258 Å) | | Cite: | Mechanism of B-box 2 domain-mediated higher-order assembly of the retroviral restriction factor TRIM5 alpha.
Elife, 5, 2016
|
|
7KM5
 
 | | Crystal structure of SARS-CoV-2 RBD complexed with Nanosota-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Spike protein S1, ... | | Authors: | Ye, G, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The development of Nanosota - 1 as anti-SARS-CoV-2 nanobody drug candidates.
Elife, 10, 2021
|
|
7KZH
 
 | |
3JCO
 
 | | Structure of yeast 26S proteasome in M1 state derived from Titan dataset | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Luan, B, Huang, X.L, Wu, J.P, Shi, Y.G, Wang, F. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of an endogenous yeast 26S proteasome reveals two major conformational states.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6Z40
 
 | |
5XZW
 
 | | Crystal structure of Rad53 1-466 | | Descriptor: | Serine/threonine-protein kinase RAD53 | | Authors: | Weng, J.H, Tsai, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phospho-Priming Confers Functionally Relevant Specificities for Rad53 Kinase Autophosphorylation
Biochemistry, 56, 2017
|
|
7MK7
 
 | | Augmentor domain of augmentor-beta | | Descriptor: | ALK and LTK ligand 1,Maltodextrin-binding protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Krimmer, S.G, Reshetnyak, A.V, Puleo, D.E, Schlessinger, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42815185 Å) | | Cite: | Structural basis for ligand reception by anaplastic lymphoma kinase.
Nature, 600, 2021
|
|
4LUQ
 
 | |
