5CNB
 
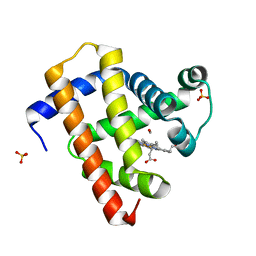 | | Ultrafast dynamics in myoglobin: 0.5 ps time delay | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
5MBM
 
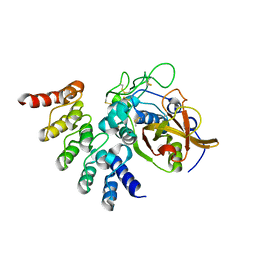 | |
1NPU
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF MURINE PD-1 | | Descriptor: | Programmed cell death protein 1 | | Authors: | Zhang, X, Schwartz, J.-C.D, Guo, X, Cao, E, Chen, L, Zhang, Z.-Y, Nathenson, S.G, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-20 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the costimulatory receptor programmed death-1.
Immunity, 20, 2004
|
|
1ZD0
 
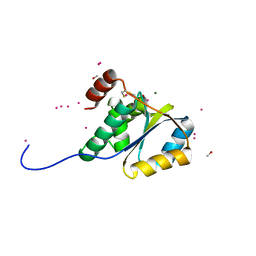 | | Crystal structure of Pfu-542154 conserved hypothetical protein | | Descriptor: | MAGNESIUM ION, METHANOL, UNKNOWN ATOM OR ION, ... | | Authors: | Habel, J.E, Liu, Z.J, Horanyi, P.S, Florence, Q.J.T, Tempel, W, Zhou, W, Chen, L, Lee, D, Nguyen, J, Chang, S.H, Bereton, P, Izumi, M, Jenny Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-04-13 | | Release date: | 2005-05-17 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Pfu-542154 conserved hypothetical protein
To be Published
|
|
7RLO
 
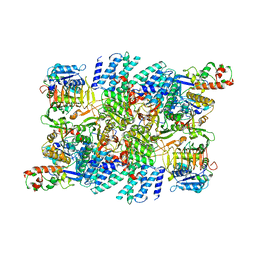 | | Structure of the human eukaryotic translation initiation factor 2B (eIF2B) in complex with a viral protein NSs | | Descriptor: | Non-structural protein NS-S, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | Authors: | Wang, L, Schoof, M, Cogan, J, Lawrence, R, Boone, M, Wuerth, J, Frost, M, Walter, P. | | Deposit date: | 2021-07-26 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Viral evasion of the integrated stress response through antagonism of eIF2-P binding to eIF2B.
Nat Commun, 12, 2021
|
|
6GEU
 
 | | Crystal structure of the C230A mutant of human IBA57 | | Descriptor: | Putative transferase CAF17, mitochondrial | | Authors: | Calderone, V, Ciofi-Baffoni, S, Gourdoupis, S, Banci, L, Nasta, V. | | Deposit date: | 2018-04-27 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | IBA57 Recruits ISCA2 to Form a [2Fe-2S] Cluster-Mediated Complex.
J.Am.Chem.Soc., 140, 2018
|
|
5YWC
 
 | |
5MF2
 
 | | Bacteriophage T5 distal tail protein pb9 co-crystallized with Tb-Xo4 | | Descriptor: | CHLORIDE ION, Distal tail protein, TERBIUM(III) ION, ... | | Authors: | Engilberge, S, Riobe, F, Di Pietro, S, Lassalle, L, Arnaud, C.-A, Breyton, C, Madern, D, Coquelle, N, Maury, O, Girard, E. | | Deposit date: | 2016-11-17 | | Release date: | 2017-09-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallophore: a versatile lanthanide complex for protein crystallography combining nucleating effects, phasing properties, and luminescence.
Chem Sci, 8, 2017
|
|
7R9W
 
 | | LC3A in complex with Fragment 1-1 | | Descriptor: | 4-phenoxybenzoic acid, GLYCEROL, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Rouge, L, Steffek, M, Helgason, E, Dueber, E, Mulvihill, M. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Multifaceted Hit-Finding Approach Reveals Novel LC3 Family Ligands.
Biochemistry, 62, 2023
|
|
7R9Z
 
 | | LC3A in complex with Fragment 2-3 | | Descriptor: | (5-fluoro-1H-indol-3-yl)acetic acid, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Rouge, L, Steffek, M, Helgason, E, Dueber, E, Mulvihill, M. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A Multifaceted Hit-Finding Approach Reveals Novel LC3 Family Ligands.
Biochemistry, 62, 2023
|
|
5MHD
 
 | |
5YW9
 
 | |
5CF1
 
 | |
1XV5
 
 | | alpha-glucosyltransferase (AGT) in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA alpha-glucosyltransferase, ... | | Authors: | Lariviere, L, Sommer, N, Morera, S. | | Deposit date: | 2004-10-27 | | Release date: | 2005-08-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural evidence of a passive base-flipping mechanism for AGT, an unusual GT-B glycosyltransferase.
J.Mol.Biol., 352, 2005
|
|
5YV7
 
 | | Racemic X-ray Structure of Calcicludine | | Descriptor: | Kunitz-type serine protease inhibitor homolog calcicludine | | Authors: | Shuai, G, Qian, Q, Lei, L. | | Deposit date: | 2017-11-24 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Racemic X-ray Structure of Calcicludine
To Be Published
|
|
7RA0
 
 | | LC3A in complex with Fragment 2-10 | | Descriptor: | (5-ethyl-2-methyl-1H-indol-3-yl)acetic acid, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Rouge, L, Steffek, M, Helgason, E, Dueber, E, Mulvihill, M. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A Multifaceted Hit-Finding Approach Reveals Novel LC3 Family Ligands.
Biochemistry, 62, 2023
|
|
1XW8
 
 | | X-ray structure of putative lactam utilization protein YBGL. Northeast Structural Genomics Consortium target ET90. | | Descriptor: | UPF0271 protein ybgL | | Authors: | Kuzin, A.P, Chen, Y, Vorobiev, S.M, Acton, T.B, Ma, L.-C, Xiao, R, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-29 | | Release date: | 2004-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of putative lactam utilization protein YBGL.
Northeast Structural Genomics Consortium target ET90.
To be Published
|
|
5MMQ
 
 | | ABA RECEPTOR FROM CITRUS, CSPYL1 | | Descriptor: | CSPYL1 | | Authors: | Moreno-Alvero, M, Yunta, C, Gonzalez-Guzman, M, Arbona, V, Granell, A, Martinez-Ripoll, M, Infantes, L, Rodriguez, P.L. | | Deposit date: | 2016-12-12 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor.
Mol Plant, 10, 2017
|
|
5Z4D
 
 | | Structure of Tailor in complex with AGUU RNA | | Descriptor: | RNA (5'-R(*AP*GP*UP*U)-3'), Terminal uridylyltransferase Tailor | | Authors: | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | Deposit date: | 2018-01-11 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
5MN0
 
 | | ABA RECEPTOR FROM CITRUS, CSPYL1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, CHLORIDE ION, CSPYL1, ... | | Authors: | Moreno-Alvero, M, Yunta, C, Gonzalez-Guzman, M, Arbona, V, Granell, A, Martinez-Ripoll, M, Infantes, L, Rodriguez, P.L, Albert, A. | | Deposit date: | 2016-12-12 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor.
Mol Plant, 10, 2017
|
|
5YWB
 
 | |
1XY9
 
 | | NMR strcutre of sst1-selective somatostatin (SRIF) analog 1 | | Descriptor: | SST1-selective somatosatin analog | | Authors: | Grace, C.R.R, Durrer, L, Koerber, S.C, Erchegyi, J, Reubi, J.C, Rivier, J.E, Riek, R. | | Deposit date: | 2004-11-09 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Somatostatin receptor 1 selective analogues: 4. Three-dimensional consensus structure by NMR
J.Med.Chem., 48, 2005
|
|
5CLR
 
 | | Crystal structure of LegK4_APO Kinase | | Descriptor: | LegK4 | | Authors: | Flayhan, A, Terradot, L. | | Deposit date: | 2015-07-16 | | Release date: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (3.706 Å) | | Cite: | The structure of Legionella pneumophila LegK4 type four secretion system (T4SS) effector reveals a novel dimeric eukaryotic-like kinase.
Sci Rep, 5, 2015
|
|
5ZBA
 
 | | Crystal structure of Rtt109-Asf1-H3-H4-CoA complex | | Descriptor: | COENZYME A, DNA damage response protein Rtt109, putative, ... | | Authors: | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | Deposit date: | 2018-02-10 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
1MZZ
 
 | | Crystal Structure of Mutant (M182T)of Nitrite Reductase | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Guo, H, Olesen, K, Xue, Y, Shapliegh, J, Sjolin, L. | | Deposit date: | 2002-10-10 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The High resolution Crystal Structures of Nitrite Reductase and its mutant Met182Thr from Rhodobacter Sphaeroides Reveal a Gating Mechanism for the Electron Transfer to the Type 1 Copper Center
To be Published
|
|
