1QGP
 
 | | NMR STRUCTURE OF THE Z-ALPHA DOMAIN OF ADAR1, 15 STRUCTURES | | Descriptor: | PROTEIN (DOUBLE STRANDED RNA ADENOSINE DEAMINASE) | | Authors: | Schade, M, Turner, C.J, Kuehne, R, Schmieder, P, Lowenhaupt, K, Herbert, A, Rich, A, Oschkinat, H. | | Deposit date: | 1999-05-03 | | Release date: | 1999-10-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Zalpha domain of the human RNA editing enzyme ADAR1 reveals a prepositioned binding surface for Z-DNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QJ7
 
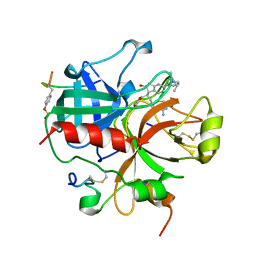 | | Novel Covalent Active Site Thrombin Inhibitors | | Descriptor: | 6-CARBAMIMIDOYL-2-[5-(3-DIETHYLCARBAMOYL-PHENYL)-2-HYDROXY-INDAN-1-YL]-HEXANOIC ACID, HIRUGEN, THROMBIN | | Authors: | Jhoti, H, Cleasby, A. | | Deposit date: | 1999-06-22 | | Release date: | 2000-06-22 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Thrombin Complexed to a Novel Series of Synthetic Inhibitors Containing a 5,5-Trans-Lactone Template
Biochemistry, 38, 1999
|
|
6H1F
 
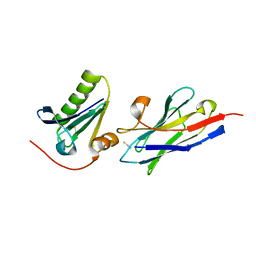 | | Structure of the nanobody-stabilized gelsolin D187N variant (second domain) | | Descriptor: | Gelsolin, THIOCYANATE ION, gelsolin nanobody, ... | | Authors: | Hassan, A, Milani, M, Mastrangelo, E, de Rosa, M. | | Deposit date: | 2018-07-11 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nanobody interaction unveils structure, dynamics and proteotoxicity of the Finnish-type amyloidogenic gelsolin variant.
Biochim Biophys Acta Mol Basis Dis, 1865, 2019
|
|
4ZBV
 
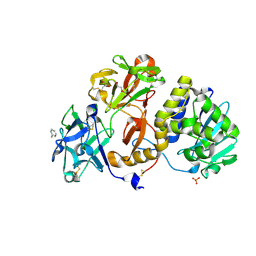 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd) in complex with benzyl T-antigen | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-15 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
1QDM
 
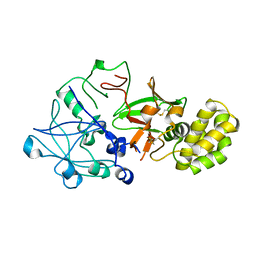 | | CRYSTAL STRUCTURE OF PROPHYTEPSIN, A ZYMOGEN OF A BARLEY VACUOLAR ASPARTIC PROTEINASE. | | Descriptor: | PROPHYTEPSIN | | Authors: | Kervinen, J, Tobin, G.J, Costa, J, Waugh, D.S, Wlodawer, A, Zdanov, A. | | Deposit date: | 1999-05-19 | | Release date: | 1999-07-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of plant aspartic proteinase prophytepsin: inactivation and vacuolar targeting.
EMBO J., 18, 1999
|
|
2PAS
 
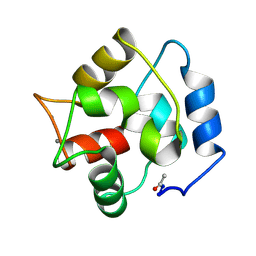 | | COMPARISON BETWEEN THE CRYSTAL AND THE SOLUTION STRUCTURES OF THE EF HAND PARVALBUMIN | | Descriptor: | CALCIUM ION, PARVALBUMIN | | Authors: | Padilla, A, Cave, A, Parello, J, Etienne, G, Baldellon, C. | | Deposit date: | 1994-03-22 | | Release date: | 1994-06-22 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Comparison between the Crystal and the Solution Structures of the EF Hand Parvalbumin
To be Published
|
|
1A70
 
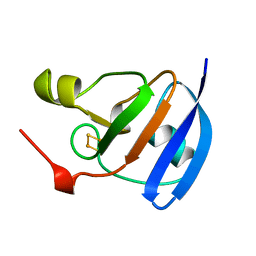 | | SPINACH FERREDOXIN | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Binda, C, Coda, A, Mattevi, A, Aliverti, A, Zanetti, G. | | Deposit date: | 1998-03-19 | | Release date: | 1998-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the mutant E92K of [2Fe-2S] ferredoxin I from Spinacia oleracea at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2KYR
 
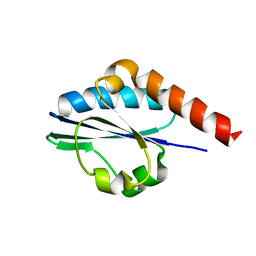 | | Solution structure of Enzyme IIB subunit of PTS system from Escherichia coli K12. Northeast Structural Genomics Consortium target ER315/Ontario Center for Structural Proteomics target ec0544 | | Descriptor: | Fructose-like phosphotransferase enzyme IIB component 1 | | Authors: | Wu, B, Yee, A, Gutmanas, A, Semest, A, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2010-06-07 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Enzyme IIB subunit of PTS system from Escherichia coli K12, Northeast Structural Genomics Consortium target ER315/Ontario Center for Structural Proteomics target ec0544
To be Published
|
|
2KZ6
 
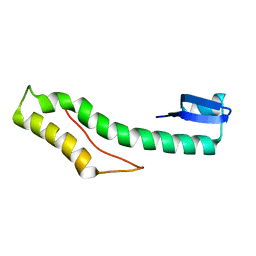 | | Solution structure of protein CV0426 from Chromobacterium violaceum, Northeast structural genomics consortium (NESG) target CVT2 | | Descriptor: | Uncharacterized protein | | Authors: | Lemak, A, Yee, A, Lee, H, Semesi, A, Prestegard, J, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-11 | | Release date: | 2010-06-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein CV0426 from Chromobacterium violaceum
To be Published
|
|
3GOY
 
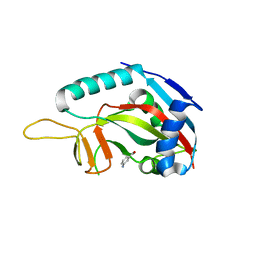 | | Crystal structure of human poly(adp-ribose) polymerase 14, catalytic fragment in complex with an inhibitor 3-aminobenzamide | | Descriptor: | 3-aminobenzamide, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Moche, M, Lehtio, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Schutz, P, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-20 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors.
Nat.Biotechnol., 30, 2012
|
|
3L3I
 
 | | Crystal structure of HLA-B*4402 in complex with the F7A mutant of a self-peptide derived from DPA*0201 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Theodossis, A, Ely, L.K, Rossjohn, J. | | Deposit date: | 2009-12-17 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Constraints within major histocompatibility complex class I restricted peptides: presentation and consequences for T-cell recognition
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3L3G
 
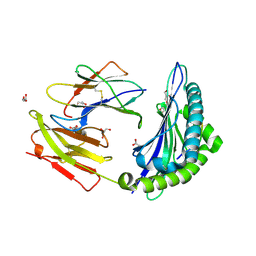 | | Crystal structure of HLA-B*4402 in complex with the R5A mutant of a self-peptide derived from DPA*0201 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Theodossis, A, Ely, L.K, Rossjohn, J. | | Deposit date: | 2009-12-16 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Constraints within major histocompatibility complex class I restricted peptides: presentation and consequences for T-cell recognition
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4E3R
 
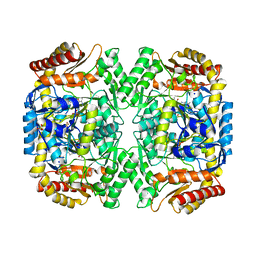 | | PLP-bound aminotransferase mutant crystal structure from Vibrio fluvialis | | Descriptor: | Pyruvate transaminase, SODIUM ION, SULFATE ION | | Authors: | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | Deposit date: | 2012-03-10 | | Release date: | 2012-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
1QLT
 
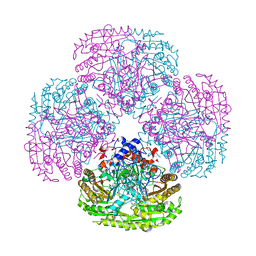 | |
5VEB
 
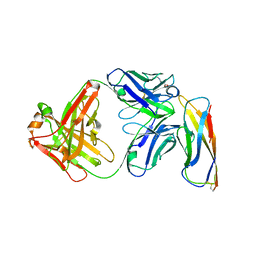 | | Crystal structure of a Fab binding to extracellular domain 5 of Cadherin-6 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Cadherin-6, anti-CDH6 Fab heavy chain, ... | | Authors: | Zhu, X, Bialucha, C.U, London, A, Clark, K, Hu, T. | | Deposit date: | 2017-04-04 | | Release date: | 2017-06-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Discovery and Optimization of HKT288, a Cadherin-6-Targeting ADC for the Treatment of Ovarian and Renal Cancers.
Cancer Discov, 7, 2017
|
|
2Q9M
 
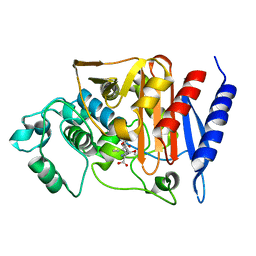 | | 4-Substituted Trinems as Broad Spectrum-Lactamase Inhibitors: Structure-based Design, Synthesis and Biological Activity | | Descriptor: | (1R,4S,7AS)-1-(1-FORMYLPROP-1-EN-1-YL)-4-METHOXY-2,4,5,6,7,7A-HEXAHYDRO-1H-ISOINDOLE-3-CARBOXYLIC ACID, Beta-lactamase | | Authors: | Plantan, I, Selic, L, Mesar, T, Stefanic Anderluh, P, Oblak, M, Prezelj, A, Hesse, L, Andrejasic, M, Vilar, M, Turk, D, Kocijan, A, Prevec, T, Vilfan, G, Kocjan, D, Copar, A, Urleb, U, Solmajer, T. | | Deposit date: | 2007-06-13 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 4-Substituted Trinems as Broad Spectrum beta-Lactamase Inhibitors: Structure-Based Design, Synthesis, and Biological Activity
J.Med.Chem., 50, 2007
|
|
5VF5
 
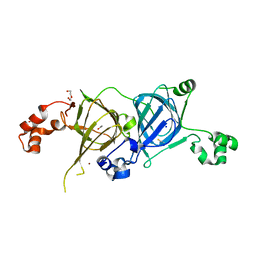 | | Crystal structure of the vicilin from Solanum melongena, re-refinement | | Descriptor: | ACETATE ION, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Porebski, P.J, Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
4Z8S
 
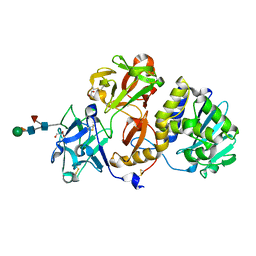 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd)-Native-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-09 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
4AYO
 
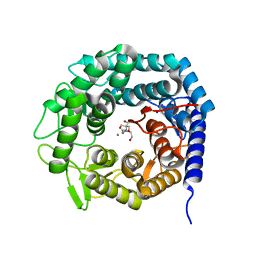 | | Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, MANNOSYL-OLIGOSACCHARIDE 1,2-ALPHA-MANNOSIDASE, ... | | Authors: | Thompson, A.J, Dabin, J, Iglesias-Fernandez, J, Iglesias-Fernandez, A, Dinev, Z, Williams, S.J, Siriwardena, A, Moreland, C, Hu, T.C, Smith, D.K, Gilbert, H.J, Rovira, C, Davies, G.J. | | Deposit date: | 2012-06-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | The Reaction Coordinate of a Bacterial Gh47 Alpha-Mannosidase: A Combined Quantum Mechanical and Structural Approach.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
1QDL
 
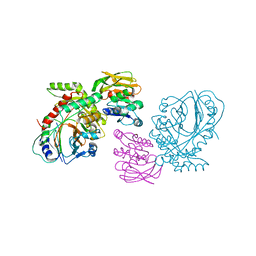 | | THE CRYSTAL STRUCTURE OF ANTHRANILATE SYNTHASE FROM SULFOLOBUS SOLFATARICUS | | Descriptor: | PROTEIN (ANTHRANILATE SYNTHASE (TRPE-SUBUNIT)), PROTEIN (ANTHRANILATE SYNTHASE (TRPG-SUBUNIT)) | | Authors: | Knoechel, T, Ivens, A, Hester, G, Gonzalez, A, Bauerle, R, Wilmanns, M, Kirschner, K, Jansonius, J.N. | | Deposit date: | 1999-05-20 | | Release date: | 1999-08-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of anthranilate synthase from Sulfolobus solfataricus: functional implications.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5VB9
 
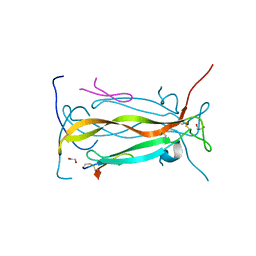 | | IL-17A in complex with peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Interleukin-17A, ... | | Authors: | Antonysamy, S, Russell, M, Zhang, A, Groshong, C, Manglicmot, D, Lu, F, Benach, J, Wasserman, S.R, Zhang, F, Afshar, S, Bina, H, Broughton, H, Chalmers, M, Dodge, J, Espada, A, Jones, S, Ting, J.P, Woodman, M. | | Deposit date: | 2017-03-28 | | Release date: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Utilization of peptide phage display to investigate hotspots on IL-17A and what it means for drug discovery.
PLoS ONE, 13, 2018
|
|
7C4O
 
 | | Solution structure of the Orange domain from human protein HES1 | | Descriptor: | Transcription factor HES-1 | | Authors: | Fan, J.S, Nayak, A, Swaminathan, K. | | Deposit date: | 2020-05-18 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Induction of Transcriptional Inhibitor Hairy and Enhancer of Split Homolog-1 and the Related Repression of Tumor-Suppressor Thioredoxin-Interacting Protein Are Important Components of Cell-Transformation Program Imposed by Oncogenic Kinase Nucleophosmin-Anaplastic Lymphoma Kinase.
Am J Pathol, 2022
|
|
3HMK
 
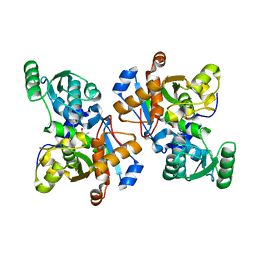 | | Crystal Structure of Serine Racemase | | Descriptor: | MANGANESE (II) ION, PYRIDOXAL-5'-PHOSPHATE, Serine racemase | | Authors: | Smith, M.A, Barker, J, Mack, V, Ebneth, A, Felicetti, B, Woods, M. | | Deposit date: | 2009-05-29 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of mammalian serine racemase: evidence for conformational changes upon inhibitor binding.
J.Biol.Chem., 285, 2010
|
|
5V4F
 
 | | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfB from Yersinia pestis | | Descriptor: | GLYCEROL, Putative translational inhibitor protein | | Authors: | Kim, Y, Chhor, G, Endres, M, Krishnan, A, Babnigg, G, Schneewind, O, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-03-09 | | Release date: | 2017-04-05 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfB from Yersinia pestis
To Be Published
|
|
5UZ7
 
 | | Volta phase plate cryo-electron microscopy structure of a calcitonin receptor-heterotrimeric Gs protein complex | | Descriptor: | Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liang, Y.L, Khoshouei, M, Radjainia, M, Zhang, Y, Glukhova, A, Tarrasch, J, Thal, D.M, Furness, S.G.B, Christopoulos, G, Coudrat, T, Danev, R, Baumeister, W, Miller, L.J, Christopoulos, A, Kobilka, B.K, Wootten, D, Skiniotis, G, Sexton, P.M. | | Deposit date: | 2017-02-24 | | Release date: | 2017-05-03 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Phase-plate cryo-EM structure of a class B GPCR-G-protein complex.
Nature, 546, 2017
|
|
