5C5S
 
 | |
8K98
 
 | | Cryo-EM structure of DSR2-TTP | | Descriptor: | a protein | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
8K9A
 
 | | Cryo-EM structure of DSR2-DSAD1 state 2 | | Descriptor: | SIR2-like domain-containing protein, SPbeta prophage-derived uncharacterized protein YotI | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
7X6Z
 
 | |
7X70
 
 | |
7X6Y
 
 | |
8IN8
 
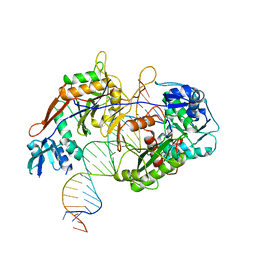 | | Cryo-EM structure of the target ssDNA-bound SIR2-APAZ/Ago-gRNA quaternary complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*GP*AP*CP*GP*TP*CP*TP*AP*AP*GP*AP*AP*AP*CP*CP*AP*TP*TP*AP*A)-3'), MAGNESIUM ION, Piwi domain protein, ... | | Authors: | Zhang, H, Li, Z, Yu, G.M, Li, X.Z, Wang, X.S. | | Deposit date: | 2023-03-08 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into mechanisms of Argonaute protein-associated NADase activation in bacterial immunity.
Cell Res., 33, 2023
|
|
8HR2
 
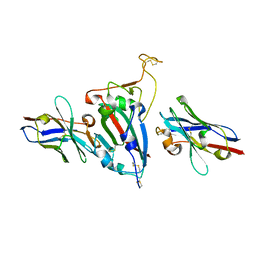 | | Ternary Crystal Complex Structure of RBD with NB1B5 and NB1C6 | | Descriptor: | NB1B5, NB1C6, Spike protein S1 | | Authors: | Sun, Z. | | Deposit date: | 2022-12-14 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure basis of two nanobodies neutralizing SARS-CoV-2 Omicron variant by targeting ultra-conservative epitopes.
J.Struct.Biol., 215, 2023
|
|
6A7A
 
 | | AKR1C1 complexed with new inhibitor with novel scaffold | | Descriptor: | (4R)-6-amino-4-(4-hydroxy-3-methoxy-5-nitrophenyl)-3-propyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X, Zhao, Y, Zhang, H, Chen, Y. | | Deposit date: | 2018-07-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
3UBU
 
 | | Crystal structure of agkisacucetin, a GpIb-binding snaclec (snake C-type lectin) that inhibits platelet | | Descriptor: | Agglucetin subunit alpha-1, Agglucetin subunit beta-2, GLYCEROL, ... | | Authors: | Gao, Y, Ge, H, Chen, H, Li, H, Liu, Y, Niu, L, Teng, M. | | Deposit date: | 2011-10-25 | | Release date: | 2012-04-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of agkisacucetin, a Gpib-binding snake C-type lectin that inhibits platelet adhesion and aggregation.
Proteins, 2012
|
|
5YYL
 
 | | Structure of Major Royal Jelly Protein 1 Oligomer | | Descriptor: | (3beta,14beta,17alpha)-ergosta-5,24(28)-dien-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tian, W, Chen, Z. | | Deposit date: | 2017-12-10 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Architecture of the native major royal jelly protein 1 oligomer.
Nat Commun, 9, 2018
|
|
2GSG
 
 | | Crystal structure of the Fv fragment of a monoclonal antibody specific for poly-glutamine | | Descriptor: | SULFATE ION, monoclonal antibody heavy chain, monoclonal antibody light chain | | Authors: | Li, P, Huey-Tubman, K.E, West Jr, A.P, Bennett, M.J, Bjorkman, P.J. | | Deposit date: | 2006-04-26 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of a polyQ-anti-polyQ complex reveals binding according to a linear lattice model.
Nat.Struct.Mol.Biol., 14, 2007
|
|
6LZ3
 
 | |
6CKH
 
 | | Manduca sexta Peptidoglycan Recognition Protein-1 | | Descriptor: | Peptidoglycan-recognition protein | | Authors: | Hu, Y. | | Deposit date: | 2018-02-28 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The three-dimensional structure and recognition mechanism of Manduca sexta peptidoglycan recognition protein-1.
Insect Biochem.Mol.Biol., 108, 2019
|
|
2W4K
 
 | | X-ray structure of a DAP-Kinase 2-302 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DEATH-ASSOCIATED PROTEIN KINASE 1, MAGNESIUM ION | | Authors: | De Diego, I, Kuper, J, Lehmann, F, Wilmanns, M. | | Deposit date: | 2008-11-27 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Pef/Y Substrate Recognition and Signature Motif Plays a Critical Role in Dapk-Related Kinase Activity.
Chem.Biol., 21, 2014
|
|
6ISM
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A1 mutant | | Descriptor: | Butyrophilin subfamily 3 member A1 | | Authors: | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
6ISO
 
 | | Human SIRT3 Recognizing H3K4cr | | Descriptor: | (2E)-BUT-2-ENAL, ARG-THR-LYS-GLN-THR-ALA-ARG, GLYCEROL, ... | | Authors: | Wang, Y, Hao, Q. | | Deposit date: | 2018-11-17 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Identification of 'erasers' for lysine crotonylated histone marks using a chemical proteomics approach.
Elife, 3, 2014
|
|
8DNK
 
 | | Crystal structure of human KRAS G12C covalently bound with Taiho WO2020/085493A1 compound 6 | | Descriptor: | 2-{[(5-tert-butyl-6-chloro-1H-indazol-3-yl)amino]methyl}-4-chloro-1-methyl-N-(1-propanoylazetidin-3-yl)-1H-imidazole-5-carboxamide, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Mohr, C. | | Deposit date: | 2022-07-11 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Modeling receptor flexibility in the structure-based design of KRAS G12C inhibitors.
J.Comput.Aided Mol.Des., 36, 2022
|
|
8DNJ
 
 | | Crystal structure of human KRAS G12C covalently bound with AstraZeneca WO2020/178282A1 compound 76 | | Descriptor: | 1-[(5S,9P,12aR)-9-(2-chloro-6-hydroxyphenyl)-8-ethynyl-10-fluoro-3,4,12,12a-tetrahydro-6H-pyrazino[2,1-c][1,4]benzoxazepin-2(1H)-yl]propan-1-one, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Mohr, C. | | Deposit date: | 2022-07-11 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Modeling receptor flexibility in the structure-based design of KRAS G12C inhibitors.
J.Comput.Aided Mol.Des., 36, 2022
|
|
8DNI
 
 | | Crystal structure of human KRAS G12C covalently bound with Araxes WO2020/028706A1 compound I-1 | | Descriptor: | (4P)-4-(5-methyl-1H-indazol-4-yl)-6-(2-propanoyl-2,6-diazaspiro[3.4]octan-6-yl)-2-(pyrrolidin-1-yl)pyrimidine-5-carbonitrile, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mohr, C. | | Deposit date: | 2022-07-11 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Modeling receptor flexibility in the structure-based design of KRAS G12C inhibitors.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5YR0
 
 | | Structure of Beclin1-UVRAG coiled coil domain complex | | Descriptor: | Beclin-1, UV radiation resistance associated protein | | Authors: | Pan, X, Zhao, Y, He, Y. | | Deposit date: | 2017-11-08 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting the potent Beclin 1-UVRAG coiled-coil interaction with designed peptides enhances autophagy and endolysosomal trafficking.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6J0L
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A3 mutant in complex with sulfate ion | | Descriptor: | Butyrophilin subfamily 3 member A3, SULFATE ION | | Authors: | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
6J0K
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A3 mutant in complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, Butyrophilin subfamily 3 member A3 | | Authors: | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
6J0G
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A3 mutant in complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, Butyrophilin subfamily 3 member A3 | | Authors: | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
6JK8
 
 | | Cryo-EM structure of the full-length human IGF-1R in complex with insulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin, ... | | Authors: | Zhang, X, Yu, D, Wang, T. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Visualization of Ligand-Bound Ectodomain Assembly in the Full-Length Human IGF-1 Receptor by Cryo-EM Single-Particle Analysis.
Structure, 28, 2020
|
|
