6ABO
 
 | | human XRCC4 and IFFO1 complex | | Descriptor: | DNA repair protein XRCC4, GLYCEROL, Intermediate filament family orphan 1, ... | | Authors: | Li, J, Liu, L, Liang, H, Liu, Y, Xu, D. | | Deposit date: | 2018-07-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The nucleoskeleton protein IFFO1 immobilizes broken DNA and suppresses chromosome translocation during tumorigenesis.
Nat.Cell Biol., 21, 2019
|
|
8XS3
 
 | | Structure of MPXV B6 and D68 fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D68_heavy chain, ... | | Authors: | wu, L.L, Sun, J.Q. | | Deposit date: | 2024-01-08 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Two noncompeting human neutralizing antibodies targeting MPXV B6 show protective effects against orthopoxvirus infections.
Nat Commun, 15, 2024
|
|
6AHV
 
 | | Crystal structure of human RPP40 | | Descriptor: | Ribonuclease P protein subunit p40 | | Authors: | Wu, J, Niu, S, Tan, M, Lan, P, Lei, M. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cryo-EM Structure of the Human Ribonuclease P Holoenzyme.
Cell, 175, 2018
|
|
8Y65
 
 | | Cryo-EM structure of human urate transporter GLUT9 bound to substrate urate | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 9, URIC ACID | | Authors: | Pan, X.J, Shen, Z.L, Xu, L, Huang, G.X.Y. | | Deposit date: | 2024-02-01 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural basis for urate recognition and apigenin inhibition of human GLUT9.
Nat Commun, 15, 2024
|
|
8Y66
 
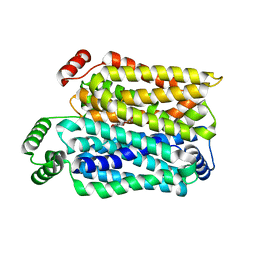 | | Cryo-EM structure of human urate transporter GLUT9 bound to inhibitor apigenin | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Solute carrier family 2, facilitated glucose transporter member 9 | | Authors: | Pan, X.J, Shen, Z.L, Xu, L, Huang, G.X.Y. | | Deposit date: | 2024-02-01 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for urate recognition and apigenin inhibition of human GLUT9.
Nat Commun, 15, 2024
|
|
8Z27
 
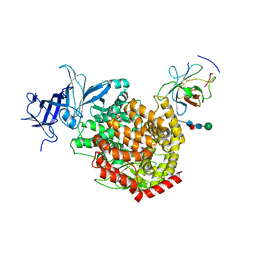 | | The structure of TGEV RBD and dog APN complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Sun, J.Q, Niu, S. | | Deposit date: | 2024-04-12 | | Release date: | 2024-12-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Cross-species recognition of two porcine coronaviruses to their cellular receptor aminopeptidase N of dogs and seven other species.
Plos Pathog., 21, 2025
|
|
8YFK
 
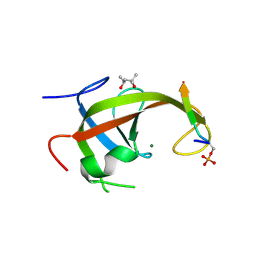 | | Crystal structure of FIP200 claw/TNIP1_FIR_pS123 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, RB1-inducible coiled-coil protein 1, ... | | Authors: | Lv, M.Q, Wu, S.M. | | Deposit date: | 2024-02-24 | | Release date: | 2024-08-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for TNIP1 binding to FIP200 during mitophagy.
J.Biol.Chem., 300, 2024
|
|
8YFM
 
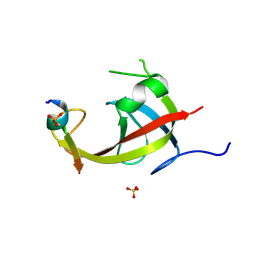 | | Crystal structure of FIP200 claw/TNIP1_FIR_pS122 | | Descriptor: | GLYCEROL, RB1-inducible coiled-coil protein 1, SULFATE ION, ... | | Authors: | Lv, M.Q, Wu, S.M. | | Deposit date: | 2024-02-24 | | Release date: | 2024-08-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for TNIP1 binding to FIP200 during mitophagy.
J.Biol.Chem., 300, 2024
|
|
8YFN
 
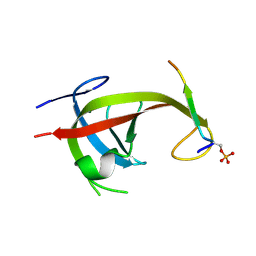 | |
8YFL
 
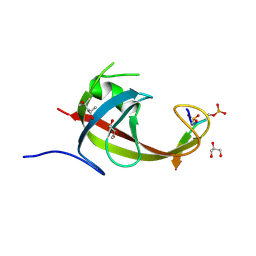 | | crystal structure of FIP200 claw/TNIP1_FIR_pS122pS123 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, RB1-inducible coiled-coil protein 1, ... | | Authors: | Lv, M.Q, Wu, S.M. | | Deposit date: | 2024-02-24 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for TNIP1 binding to FIP200 during mitophagy.
J.Biol.Chem., 300, 2024
|
|
8YZI
 
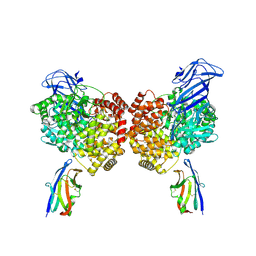 | | The structure of PDCoV RBD and dog APN complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Sun, J.Q, Niu, S. | | Deposit date: | 2024-04-07 | | Release date: | 2024-12-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cross-species recognition of two porcine coronaviruses to their cellular receptor aminopeptidase N of dogs and seven other species.
Plos Pathog., 21, 2025
|
|
7E1V
 
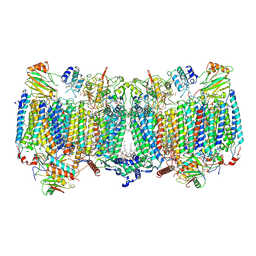 | | Cryo-EM structure of apo hybrid respiratory supercomplex consisting of Mycobacterium tuberculosis complexIII and Mycobacterium smegmatis complexIV | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, CARDIOLIPIN, ... | | Authors: | Zhou, S, Wang, W, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2021-02-03 | | Release date: | 2021-10-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structure of Mycobacterium tuberculosis cytochrome bcc in complex with Q203 and TB47, two anti-TB drug candidates.
Elife, 10, 2021
|
|
8ZW8
 
 | |
8ZXP
 
 | | Cryo-EM structure of TmaT-TMA complexes | | Descriptor: | N,N-dimethylmethanamine, Trimethylamine transporter | | Authors: | Chao, G. | | Deposit date: | 2024-06-14 | | Release date: | 2024-11-06 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis of a microbial trimethylamine transporter.
Mbio, 16, 2025
|
|
8ZXK
 
 | |
7E1X
 
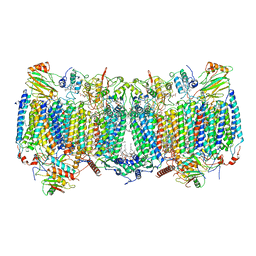 | | Cryo-EM structure of hybrid respiratory supercomplex consisting of Mycobacterium tuberculosis complexIII and Mycobacterium smegmatis complexIV in presence of TB47 | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, 5-methoxy-2-methyl-~{N}-[[4-[4-[4-(trifluoromethyloxy)phenyl]piperidin-1-yl]phenyl]methyl]pyrazolo[1,5-a]pyridine-3-carboxamide, ... | | Authors: | Zhou, S, Wang, W, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2021-02-03 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structure of Mycobacterium tuberculosis cytochrome bcc in complex with Q203 and TB47, two anti-TB drug candidates.
Elife, 10, 2021
|
|
7E1W
 
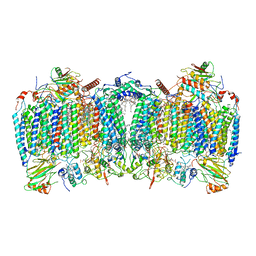 | | Cryo-EM structure of hybrid respiratory supercomplex consisting of Mycobacterium tuberculosis complexIII and Mycobacterium smegmatis complexIV in the presence of Q203 | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, 6-chloranyl-2-ethyl-N-[[4-[4-[4-(trifluoromethyloxy)phenyl]piperidin-1-yl]phenyl]methyl]imidazo[1,2-a]pyridine-3-carboxamide, ... | | Authors: | Zhou, S, Wang, W, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2021-02-03 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structure of Mycobacterium tuberculosis cytochrome bcc in complex with Q203 and TB47, two anti-TB drug candidates.
Elife, 10, 2021
|
|
6AHU
 
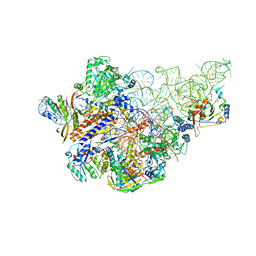 | | Cryo-EM structure of human Ribonuclease P with mature tRNA | | Descriptor: | H1 RNA, Ribonuclease P protein subunit p14, Ribonuclease P protein subunit p20, ... | | Authors: | Wu, J, Niu, S, Tan, M, Lan, P, Lei, M. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM Structure of the Human Ribonuclease P Holoenzyme.
Cell, 175, 2018
|
|
6AHR
 
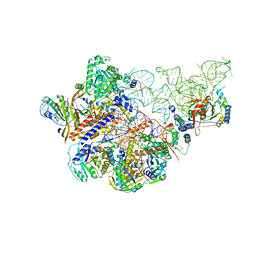 | | Cryo-EM structure of human Ribonuclease P | | Descriptor: | H1 RNA, Ribonuclease P protein subunit p14, Ribonuclease P protein subunit p20, ... | | Authors: | Wu, J, Niu, S, Tan, M, Lan, P, Lei, M. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Cryo-EM Structure of the Human Ribonuclease P Holoenzyme.
Cell, 175, 2018
|
|
6K7O
 
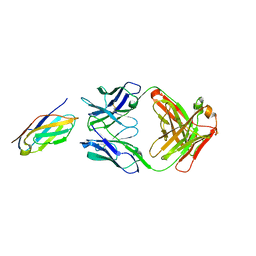 | | Complex structure of LILRB4 and h128-3 antibody | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily B member 4, h128-3 Fab heavy chain, h128-3 Fab light chain | | Authors: | Song, H, Chai, Y, Xu, X, Gao, F.G. | | Deposit date: | 2019-06-08 | | Release date: | 2019-07-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Disrupting LILRB4/APOE Interaction by an Efficacious Humanized Antibody Reverses T-cell Suppression and Blocks AML Development.
Cancer Immunol Res, 7, 2019
|
|
5ZAZ
 
 | |
7BZ5
 
 | | Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of B38, Light chain of B38, ... | | Authors: | Wu, Y, Qi, J, Gao, F. | | Deposit date: | 2020-04-26 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2.
Science, 368, 2020
|
|
7WB4
 
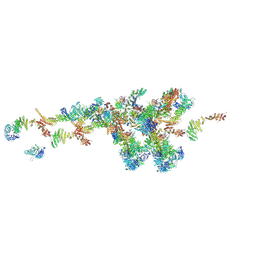 | | Cryo-EM structure of the NR subunit from X. laevis NPC | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Huang, G, Zhan, X, Shi, Y. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Cryo-EM structure of the nuclear ring from Xenopus laevis nuclear pore complex.
Cell Res., 32, 2022
|
|
8JD9
 
 | | Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Sodium/hydrogen exchanger 7 | | Authors: | Yang, G.H, Zhang, Y.M, Zhou, J.Q, Jia, Y.T, Xu, X, Fu, P, Wu, H.Y. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance.
Nat.Plants, 9, 2023
|
|
8JDA
 
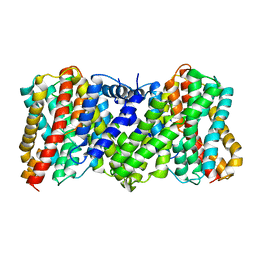 | | Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class2 | | Descriptor: | Sodium/hydrogen exchanger 7 | | Authors: | Yang, G.H, Zhang, Y.M, Zhou, J.Q, Jia, Y.T, Xu, X, Fu, P, Wu, H.Y. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance.
Nat.Plants, 9, 2023
|
|
