7ABM
 
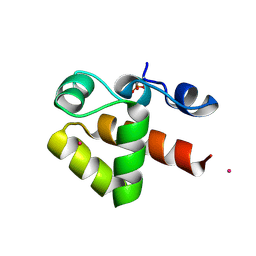 | | X-ray structure of phosphorylated Barrier-to-autointegration factor (BAF) | | Descriptor: | Barrier-to-autointegration factor, CESIUM ION | | Authors: | Zinn-Justin, S, Marcelot, A, Le Du, M.H, Ropars, V. | | Deposit date: | 2020-09-08 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Di-phosphorylated BAF shows altered structural dynamics and binding to DNA, but interacts with its nuclear envelope partners.
Nucleic Acids Res., 49, 2021
|
|
8FAT
 
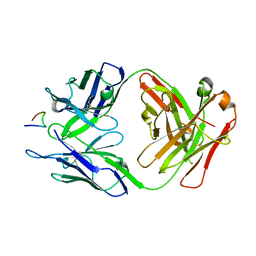 | | Crystal structure of Ky224 Fab in complex with circumsporozoite protein NPDP peptide | | Descriptor: | Circumsporozoite protein NPDP peptide, Ky224 Antibody, heavy chain, ... | | Authors: | Kassardjian, A, Thai, E, Julien, J.P. | | Deposit date: | 2022-11-28 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular determinants of cross-reactivity and potency by VH3-33 antibodies against the Plasmodium falciparum circumsporozoite protein.
Cell Rep, 42, 2023
|
|
1A4T
 
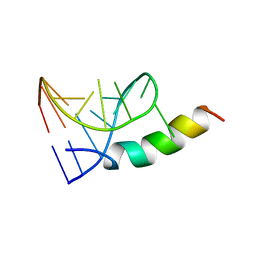 | | SOLUTION STRUCTURE OF PHAGE P22 N PEPTIDE-BOX B RNA COMPLEX, NMR, 20 STRUCTURES | | Descriptor: | 20-MER BASIC PEPTIDE, BOXB RNA | | Authors: | Cai, Z, Gorin, A.A, Frederick, R, Ye, X, Hu, W, Majumdar, A, Kettani, A, Patel, D.J. | | Deposit date: | 1998-02-04 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of P22 transcriptional antitermination N peptide-boxB RNA complex.
Nat.Struct.Biol., 5, 1998
|
|
6ZVG
 
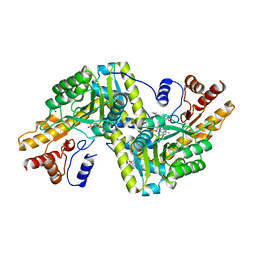 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - L-indole-3-lactic acid | | Descriptor: | 3-(INDOL-3-YL) LACTATE, Aminotransferase, MAGNESIUM ION, ... | | Authors: | Rum, J, Rutkiewicz, M, Pruska, A, Bujacz, A, Pietrzyk-Brzezinska, A.J, Bujacz, G. | | Deposit date: | 2020-07-24 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials (Basel), 14, 2021
|
|
3IRQ
 
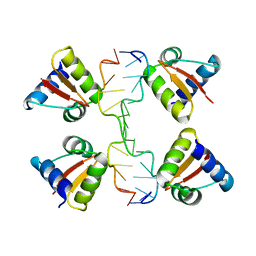 | | Crystal structure of a Z-Z junction | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*CP*GP*CP*GP*AP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*TP*CP*GP*CP*GP*CP*G)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Athanasiadis, A, de Rosa, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a junction between two Z-DNA helices.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6ZWK
 
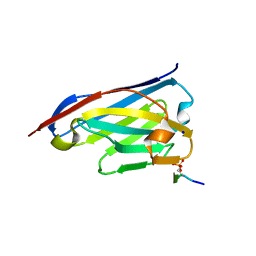 | | Crystal structure of the phosphorylated C-terminal tail of histone H2AX in complex with a specific nanobody (C6 gammaXbody) | | Descriptor: | CHLORIDE ION, Histone H2AX, SODIUM ION, ... | | Authors: | McEwen, A.G, Moeglin, E, Desplancq, D, Weiss, E, Poterszman, A. | | Deposit date: | 2020-07-28 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Novel Nanobody Precisely Visualizes Phosphorylated Histone H2AX in Living Cancer Cells under Drug-Induced Replication Stress.
Cancers (Basel), 13, 2021
|
|
8PWZ
 
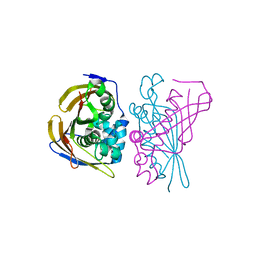 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadBD from Mycobacterium tuberculosis | | Descriptor: | (3R)-hydroxyacyl-ACP dehydratase subunit HadB, UPF0336 protein Rv0504c | | Authors: | Rima, J, Grimoire, Y, Bories, P, Bardou, F, Quemard, A, Bon, C, Mourey, L, Tranier, S. | | Deposit date: | 2023-07-22 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.00196719 Å) | | Cite: | HadBD dehydratase from Mycobacterium tuberculosis fatty acid synthase type II: A singular structure for a unique function.
Protein Sci., 33, 2024
|
|
8FAS
 
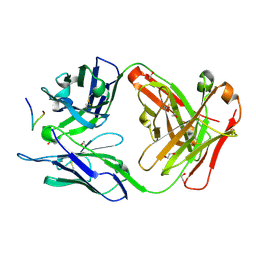 | | Crystal structure of Ky230 Fab in complex with circumsporozoite protein NANP5 peptide | | Descriptor: | 1,2-ETHANEDIOL, Circumsporozoite protein NANP5 peptide, Ky230 Antibody, ... | | Authors: | Kassardjian, A, Thai, E, Julien, J.P. | | Deposit date: | 2022-11-28 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular determinants of cross-reactivity and potency by VH3-33 antibodies against the Plasmodium falciparum circumsporozoite protein.
Cell Rep, 42, 2023
|
|
1NU0
 
 | | Structure of the double mutant (L6M; F134M, SeMet form) of yqgF from Escherichia coli, a hypothetical protein | | Descriptor: | Hypothetical protein yqgF, SULFATE ION | | Authors: | Galkin, A, Sarikaya, E, Krajewski, W, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-30 | | Release date: | 2004-03-02 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of yqgF from Escherichia coli, a hypothetical protein
To be Published
|
|
5ZYX
 
 | | Solution NMR structure of K30 peptide in 10 mM dioctanoyl phosphatidylglycerol (D8PG) | | Descriptor: | ARG-TRP-LYS-ARG-HIS-ILE-SER-GLU-GLN-LEU-ARG-ARG-ARG-ASP-ARG-LEU-GLN-ARG-GLN-ALA | | Authors: | Bhunia, A, Mohid, A, Stella, L, Calligari, P. | | Deposit date: | 2018-05-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design, Synthesis, Antibacterial Potential, and Structural Characterization of N-Acylated Derivatives of the Human Autophagy 16 Polypeptide.
Bioconjug.Chem., 30, 2019
|
|
4XI1
 
 | | Crystal structure of U-box 2 of LubX / LegU2 / Lpp2887 from Legionella pneumophila str. Paris, wild-type | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase LubX, GLYCEROL, ... | | Authors: | Stogios, P.J, Quaile, T, Skarina, T, Cuff, M, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-06 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.983 Å) | | Cite: | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
1NXD
 
 | | Crystal structure of MnMn Concanavalin A | | Descriptor: | AZIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lopez-Jaramillo, F.J, Gonzalez-Ramirez, L.A, Albert, A, Santoyo-Gonzalez, F, Vargas-Berenguel, A, Otalora, F. | | Deposit date: | 2003-02-10 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of concanavalin A at pH 8: bound solvent and crystal contacts.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6SAI
 
 | |
6SCD
 
 | | Polyester hydrolase PE-H Y250S mutant of Pseudomonas aestusnigri | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bollinger, A, Thies, S, Kobus, S, Hoeppner, A, Smits, S.H.J, Jaeger, K.-E. | | Deposit date: | 2019-07-24 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A Novel Polyester Hydrolase From the Marine BacteriumPseudomonas aestusnigri -Structural and Functional Insights.
Front Microbiol, 11, 2020
|
|
6SA7
 
 | | DARPin-Armadillo fusion C8long83 | | Descriptor: | DARPin-Armadillo fusion C8long83 | | Authors: | Ernst, P, Honegger, A, van der Valk, F, Ewald, C, Mittl, P.R.E, Plucktun, A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Rigid fusions of designed helical repeat binding proteins efficiently protect a binding surface from crystal contacts.
Sci Rep, 9, 2019
|
|
6SBN
 
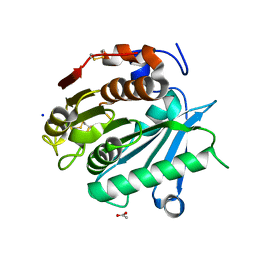 | | Polyester hydrolase PE-H of Pseudomonas aestusnigri | | Descriptor: | ACETATE ION, SODIUM ION, polyester hydrolase | | Authors: | Bollinger, A, Thies, S, Kobus, S, Hoeppner, A, Smits, S.H.J, Jaeger, K.-E. | | Deposit date: | 2019-07-22 | | Release date: | 2020-02-26 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | A Novel Polyester Hydrolase From the Marine BacteriumPseudomonas aestusnigri -Structural and Functional Insights.
Front Microbiol, 11, 2020
|
|
6EON
 
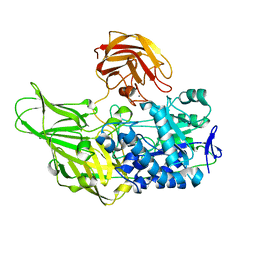 | | Galactanase BT0290 | | Descriptor: | Beta-galactosidase, CALCIUM ION, alpha-D-galactopyranose | | Authors: | Basle, A, Munoz, J, Gilbert, H. | | Deposit date: | 2017-10-10 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
3ILB
 
 | |
8G2I
 
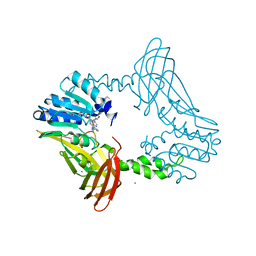 | | Crystal Structure of PRMT4 with Compound YD1290 | | Descriptor: | 5'-([2-(benzylcarbamamido)ethyl]{3-[N'-(3-bromophenyl)carbamimidamido]propyl}amino)-5'-deoxyadenosine, Histone-arginine methyltransferase CARM1, UNKNOWN ATOM OR ION | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A unique binding pocket induced by a noncanonical SAH mimic to develop potent and selective PRMT inhibitors.
Acta Pharm Sin B, 13, 2023
|
|
3IGO
 
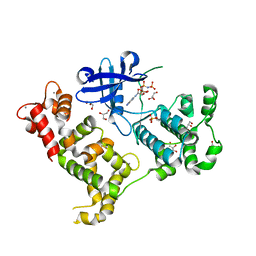 | | Crystal structure of Cryptosporidium parvum CDPK1, cgd3_920 | | Descriptor: | CALCIUM ION, Calmodulin-domain protein kinase 1, GLYCEROL, ... | | Authors: | Wernimont, A.K, Artz, J.D, Finnerty, P, Amani, M, Allali-Hassanali, A, Vedadi, M, Tempel, W, MacKenzie, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Bochkarev, A, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-28 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3RAO
 
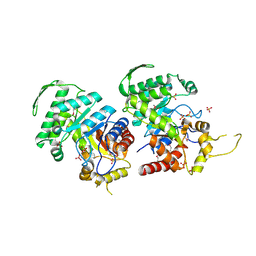 | | Crystal Structure of the Luciferase-like Monooxygenase from Bacillus cereus ATCC 10987. | | Descriptor: | Putative Luciferase-like Monooxygenase, SULFATE ION | | Authors: | Domagalski, M.J, Chruszcz, M, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-28 | | Release date: | 2011-05-11 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Luciferase-like Monooxygenase from Bacillus cereus ATCC 10987.
To be Published
|
|
7AA4
 
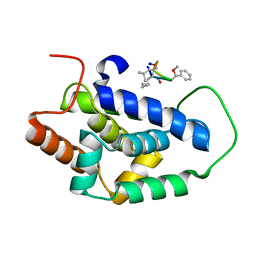 | | Structure of ClpC1-NTD bound to a CymA analogue | | Descriptor: | Negative regulator of genetic competence ClpC/mecB, polymer Cyclomarin A analogue | | Authors: | Meinhart, A, Morreale, F.E, Kaiser, M, Clausen, T. | | Deposit date: | 2020-09-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
7A3W
 
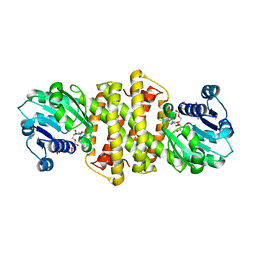 | | Structure of Imine Reductase from Pseudomonas sp. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NAD(P)-dependent oxidoreductase, ... | | Authors: | Cuetos, A, Thorpe, T, Turner, N.J, Grogan, G. | | Deposit date: | 2020-08-18 | | Release date: | 2021-08-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Multifunctional biocatalyst for conjugate reduction and reductive amination.
Nature, 604, 2022
|
|
3T7L
 
 | | Crystal structure of the FYVE domain of endofin (ZFYVE16) at 1.1A resolution | | Descriptor: | 1,2-ETHANEDIOL, ZINC ION, Zinc finger FYVE domain-containing protein 16 | | Authors: | Chaikuad, A, Williams, E, Guo, K, Sanvitale, C, Berridge, G, Krojer, T, Muniz, J.R.C, Canning, P, Phillips, C, Shrestha, A, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-07-30 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Crystal structure of the FYVE domain of endofin (ZFYVE16) at 1.1A resolution
To be Published
|
|
3IB0
 
 | | Structural basis of the prevention of NSAID-induced damage of the gastrointestinal tract by C-terminal half (C-lobe) of bovine colostrum protein lactoferrin: Binding and structural studies of C-lobe complex with diclofenac | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Mir, R, Singh, N, Sinha, M, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2009-07-15 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structural basis for the prevention of nonsteroidal antiinflammatory drug-induced gastrointestinal tract damage by the C-lobe of bovine colostrum lactoferrin
Biophys.J., 97, 2009
|
|
