6NAE
 
 | |
7CMK
 
 | | E30 E-particle in complex with 6C5 | | Descriptor: | Heavy chain, Light chain, VP1, ... | | Authors: | Wang, K, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-07-27 | | Release date: | 2020-08-12 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Serotype specific epitopes identified by neutralizing antibodies underpin immunogenic differences in Enterovirus B.
Nat Commun, 11, 2020
|
|
5VID
 
 | |
5VLI
 
 | | Computationally designed inhibitor peptide HB1.6928.2.3 in complex with influenza hemagglutinin (A/PuertoRico/8/1934) | | Descriptor: | 2,5,8,11-TETRAOXATRIDECANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bernard, S.M, Wilson, I.A. | | Deposit date: | 2017-04-25 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Massively parallel de novo protein design for targeted therapeutics.
Nature, 550, 2017
|
|
6NCU
 
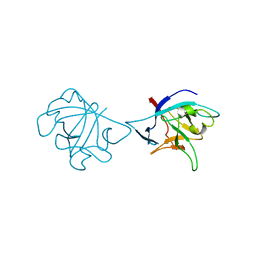 | | Interleukin-37 residues 53-206- dimer | | Descriptor: | Interleukin-37 | | Authors: | Eisenmesser, E.Z. | | Deposit date: | 2018-12-12 | | Release date: | 2019-03-13 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Interleukin-37 monomer is the active form for reducing innate immunity.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NX1
 
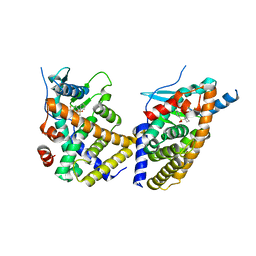 | | STRUCTURE OF HUMAN PREGNANE X RECEPTOR LIGAND BINDING DOMAIN BOUND TETHERED WITH SRC CO-ACTIVATOR PEPTIDE AND COMPOUND-3 AKA 1,1,1,3,3,3-HEXAFLUORO-2-{4-[1-(4- LUOROBENZENESULFONYL)CYCLOPENTYL]PHENYL}PROPAN-2-OL | | Descriptor: | 1,1,1,3,3,3-hexafluoro-2-(4-{1-[(4-fluorophenyl)sulfonyl]cyclopentyl}phenyl)propan-2-ol, Nuclear receptor subfamily 1 group I member 2,Nuclear receptor coactivator 1 fusion | | Authors: | Khan, J.A. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-based Discovery of Phenyl (3-Phenylpyrrolidin-3-yl)sulfones as Selective, Orally Active ROR gamma t Inverse Agonists.
Acs Med.Chem.Lett., 10, 2019
|
|
6O1S
 
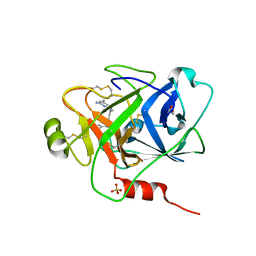 | | Structure of human plasma kallikrein protease domain with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, N-[(6-amino-2,4-dimethylpyridin-3-yl)methyl]-1-({4-[(1H-pyrazol-1-yl)methyl]phenyl}methyl)-1H-pyrazole-4-carboxamide, PHOSPHATE ION, ... | | Authors: | Partridge, J.R, Choy, R.M. | | Deposit date: | 2019-02-21 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of full-length plasma kallikrein bound to highly specific inhibitors describe a new mode of targeted inhibition.
J.Struct.Biol., 206, 2019
|
|
6O98
 
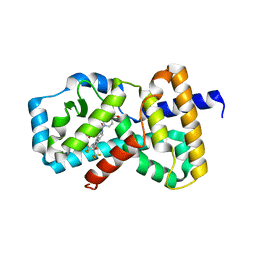 | | Crystal structure of RAR-related orphan receptor C in complex with a phenyl (3-phenylpyrrolidin-3-yl)sulfone inhibitor | | Descriptor: | 1-(4-{(3R)-3-[(4-fluorophenyl)sulfonyl]-3-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]pyrrolidine-1-carbonyl}piperazin-1-yl)ethan-1-one, Nuclear receptor ROR-gamma | | Authors: | Sack, J.S. | | Deposit date: | 2019-03-13 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-based Discovery of Phenyl (3-Phenylpyrrolidin-3-yl)sulfones as Selective, Orally Active ROR gamma t Inverse Agonists.
ACS Med Chem Lett, 10, 2019
|
|
5VMR
 
 | |
7YHK
 
 | | Cryo-EM structure of the HA trimer of A/Beijing/262/1995(H1N1) in complex with neutralizing antibody 12H5 | | Descriptor: | 12H5 heavy chain, 12H5 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zheng, Q, Li, S, Li, T, Xue, W, Sun, H. | | Deposit date: | 2022-07-13 | | Release date: | 2022-08-17 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses.
Nat Commun, 13, 2022
|
|
1CF5
 
 | | BETA-MOMORCHARIN STRUCTURE AT 2.55 A | | Descriptor: | PROTEIN (BETA-MOMORCHARIN), beta-D-xylopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yuan, Y.-R, He, Y.-N, Xiong, J.-P, Xia, Z.-X. | | Deposit date: | 1999-03-24 | | Release date: | 1999-06-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Three-dimensional structure of beta-momorcharin at 2.55 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
8IPQ
 
 | |
6OUS
 
 | | Structure of fusion glycoprotein from human respiratory syncytial virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F1 fused with Fibritin trimerization domain, Fusion glycoprotein F2, ... | | Authors: | Su, H.P. | | Deposit date: | 2019-05-05 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A potent broadly neutralizing human RSV antibody targets conserved site IV of the fusion glycoprotein.
Nat Commun, 10, 2019
|
|
8IPT
 
 | |
8IPS
 
 | |
8IPR
 
 | |
8KB0
 
 | | Crystal structure of 01JD-AEAIIVAMV | | Descriptor: | Beta2-microglobulin, MHC class I antigen alpha chain, peptide of AIV | | Authors: | Tang, Z, Zhang, N. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The impact of micropolymorphism in Anpl-UAA on structural stability and peptide presentation.
Int.J.Biol.Macromol., 267, 2024
|
|
8KB1
 
 | | Crystal structure of 11JD | | Descriptor: | Beta2-microglobulin, MHC class I antigen, peptide of AIV | | Authors: | Tang, Z, Zhang, N. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The impact of micropolymorphism in Anpl-UAA on structural stability and peptide presentation.
Int.J.Biol.Macromol., 267, 2024
|
|
7BWJ
 
 | | crystal structure of SARS-CoV-2 antibody with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | Authors: | Wang, X, Ge, J. | | Deposit date: | 2020-04-14 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Human neutralizing antibodies elicited by SARS-CoV-2 infection.
Nature, 584, 2020
|
|
8PY3
 
 | | Crystal structure of human Sirt2 in complex with a 1,2,4-oxadiazole based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-chloranyl-~{N}-[4-[5-[[(3~{S})-1-[(3-fluoranyl-2-methyl-phenyl)methyl]piperidin-3-yl]methyl]-1,2,4-oxadiazol-3-yl]phenyl]benzamide, ... | | Authors: | Friedrich, F, Colcerasa, A, Einsle, O, Jung, M. | | Deposit date: | 2023-07-24 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Activity Studies of 1,2,4-Oxadiazoles for the Inhibition of the NAD + -Dependent Lysine Deacylase Sirtuin 2.
J.Med.Chem., 67, 2024
|
|
8QJ3
 
 | |
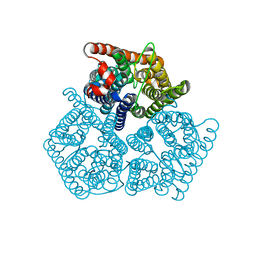
8QJ4
 
 | |
8JNC
 
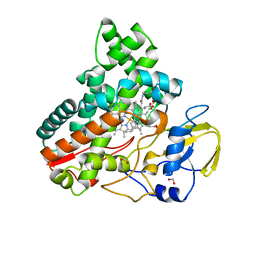 | | Crystal structure of cytochrome P450 IkaD from Streptomyces sp. ZJ306, in complex with the substrate 10-epi-maltophilin | | Descriptor: | (1Z,3E,5S,8R,9S,10S,11R,13R,15R,16S,18Z,24S,25S)-11-ethyl-2,24-dihydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.09,13.08,15.05,16]octacosa-1(2),3,18-triene-7,20,27,28-tetraone, Cytochrome P450, FORMIC ACID, ... | | Authors: | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8JNQ
 
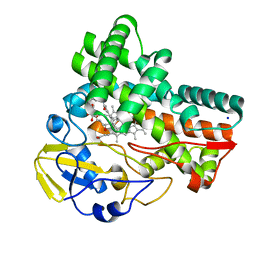 | | Crystal structure of cytochrome P450 CftA from Streptomyces torulosus NRRL B-3889, in complex with a substrate compound c | | Descriptor: | (1Z,3E,5E,7S,8R,10S,11R,13R,15R,16E,18E,25S)-11-ethyl-2,7-dihydroxy-10-methyl-21,26-diazatetracyclo[23.2.1.09,13.08,15]octacosa-1(2),3,5,16,18-pentaene-20,27,28-trione, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Jiang, P, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8JNP
 
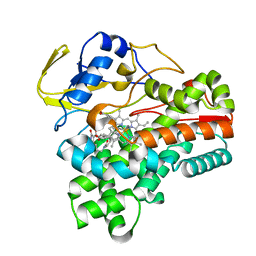 | | Crystal structure of cytochrome P450 CftA from Streptomyces torulosus NRRL B-3889, in complex with the substrate ikarugamycin | | Descriptor: | (1Z,3E,5S,7R,8R,10R,11R,12S,15R,16S,18Z,25S)-11-ethyl-2-hydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.05,16.07,15.08,12]octacosa-1(2),3,13,18-tetraene-20,27,28-trione, Cytochrome P450 CftA, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, P, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
