6LJT
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3-chloranyl-2-phenyl-phenyl)amino]benzoic acid, Fatty acid-binding protein, ... | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
6LQH
 
 | | High resolution architecture of curli complex | | Descriptor: | Curli production assembly/transport component CsgF, Curli production assembly/transport component CsgG | | Authors: | Zhang, M, Shi, H, Huang, Y. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-15 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cryo-EM structure of the nonameric CsgG-CsgF complex and its implications for controlling curli biogenesis in Enterobacteriaceae.
Plos Biol., 18, 2020
|
|
6LQJ
 
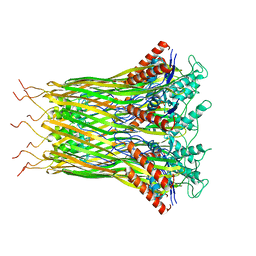 | | Low resolution architecture of curli complex | | Descriptor: | Curli production assembly/transport component CsgF, Curli production assembly/transport component CsgG | | Authors: | Zhang, M, Shi, H, Huang, Y. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Cryo-EM structure of the nonameric CsgG-CsgF complex and its implications for controlling curli biogenesis in Enterobacteriaceae.
Plos Biol., 18, 2020
|
|
5NSS
 
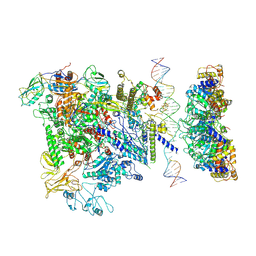 | | Cryo-EM structure of RNA polymerase-sigma54 holoenzyme with promoter DNA and transcription activator PspF intermedate complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta',DNA-directed RNA polymerase subunit beta',DNA-directed RNA polymerase subunit beta', ... | | Authors: | Glyde, R, Ye, F.Z, Darbari, V.C, Zhang, N, Buck, M, Zhang, X.D. | | Deposit date: | 2017-04-26 | | Release date: | 2017-06-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structures of RNA Polymerase Closed and Intermediate Complexes Reveal Mechanisms of DNA Opening and Transcription Initiation.
Mol. Cell, 67, 2017
|
|
4Y33
 
 | | Crystal of NO66 in complex with Ni(II)and N-oxalylglycine (NOG) | | Descriptor: | Bifunctional lysine-specific demethylase and histidyl-hydroxylase NO66, N-OXALYLGLYCINE, NICKEL (II) ION | | Authors: | Wang, C, Zhang, Q, Zang, J. | | Deposit date: | 2015-02-10 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the JmjC domain-containing protein NO66 complexed with ribosomal protein Rpl8.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5NSR
 
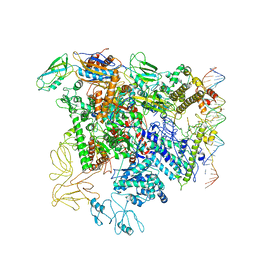 | | Cryo-EM structure of RNA polymerase-sigma54 holo enzyme with promoter DNA closed complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Glyde, R, Ye, F.Z, Darbari, V.C, Zhang, N, Buck, M, Zhang, X.D. | | Deposit date: | 2017-04-26 | | Release date: | 2017-06-28 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of RNA Polymerase Closed and Intermediate Complexes Reveal Mechanisms of DNA Opening and Transcription Initiation.
Mol. Cell, 67, 2017
|
|
4Y3O
 
 | | Crystal structure of Ribosomal oxygenase NO66 in complex with substrate Rpl8 peptide and Ni(II) and cofactor N-oxalyglycine | | Descriptor: | ACETATE ION, Bifunctional lysine-specific demethylase and histidyl-hydroxylase NO66, GLYCEROL, ... | | Authors: | Wang, C, Zhang, Q, Zang, J. | | Deposit date: | 2015-02-10 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the JmjC domain-containing protein NO66 complexed with ribosomal protein Rpl8.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1JRU
 
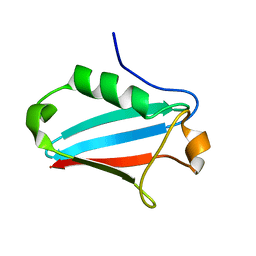 | | NMR STRUCTURE OF THE UBX DOMAIN FROM P47 (ENERGY MINIMISED AVERAGE) | | Descriptor: | p47 protein | | Authors: | Yuan, X.M, Shaw, A, Zhang, X.D, Kondo, H, Lally, J, Freemont, P.S, Matthews, S.J. | | Deposit date: | 2001-08-15 | | Release date: | 2001-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and interaction surface of the C-terminal domain from p47: a major p97-cofactor involved in SNARE disassembly.
J.Mol.Biol., 311, 2001
|
|
3NFC
 
 | |
4UT7
 
 | | CRYSTAL STRUCTURE OF THE SCFV FRAGMENT OF THE BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11 | | Descriptor: | BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11 | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
4LA2
 
 | | Crystal structure of dimethylsulphoniopropionate (DMSP) lyase DddQ | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Dimethylsulphoniopropionate (DMSP) lyase DddQ, ZINC ION | | Authors: | Zhang, Y, Li, C. | | Deposit date: | 2013-06-19 | | Release date: | 2014-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insight into bacterial cleavage of oceanic dimethylsulfoniopropionate into dimethyl sulfide
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4LA3
 
 | |
9LGM
 
 | | Cryo-EM structure of GPR4 complexed with Gs in pH8.0 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yue, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2025-01-10 | | Release date: | 2025-04-23 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis of stepwise proton sensing-mediated GPCR activation.
Cell Res., 35, 2025
|
|
6GH5
 
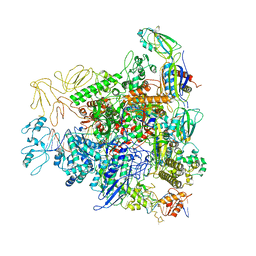 | | Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme transcription open complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Glyde, R, Ye, F.Z, Zhang, X.D. | | Deposit date: | 2018-05-04 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Bacterial RNA Polymerase Complexes Reveal the Mechanism of DNA Loading and Transcription Initiation.
Mol. Cell, 70, 2018
|
|
6GFW
 
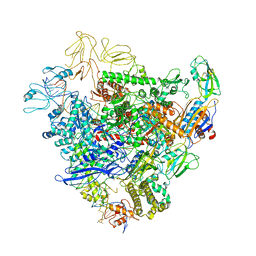 | | Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme initial transcribing complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Glyde, R, Ye, F.Z, Zhang, X.D. | | Deposit date: | 2018-05-02 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of Bacterial RNA Polymerase Complexes Reveal the Mechanism of DNA Loading and Transcription Initiation.
Mol. Cell, 70, 2018
|
|
6GH6
 
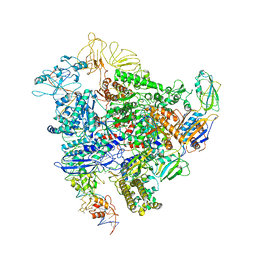 | | Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme intermediate partially loaded complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Glyde, R, Ye, F.Z, Zhang, X.D. | | Deposit date: | 2018-05-04 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of Bacterial RNA Polymerase Complexes Reveal the Mechanism of DNA Loading and Transcription Initiation.
Mol. Cell, 70, 2018
|
|
6OX2
 
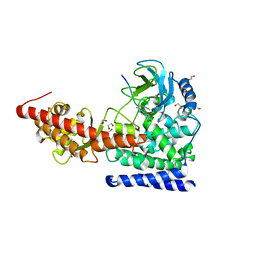 | | SETD3in Complex with an Actin Peptide with the Target Histidine Fully Methylated | | Descriptor: | 1,2-ETHANEDIOL, Actin Peptide, GLYCEROL, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2019-05-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
6L6H
 
 | | Crystal structure of Lpg0189 | | Descriptor: | ACETATE ION, GLYCEROL, Uncharacterized protein Lpg0189 | | Authors: | Ge, H, Chen, X. | | Deposit date: | 2019-10-29 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Crystal structure of a hypothetical T2SS effector Lpg0189 from Legionella pneumophila reveals a novel protein fold.
Biochem.Biophys.Res.Commun., 521, 2020
|
|
6L6G
 
 | | Crystal structure of SeMet_Lpg0189 | | Descriptor: | GLYCEROL, Uncharacterized protein Lpg0189 | | Authors: | Ge, H, Chen, X. | | Deposit date: | 2019-10-29 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of a hypothetical T2SS effector Lpg0189 from Legionella pneumophila reveals a novel protein fold.
Biochem.Biophys.Res.Commun., 521, 2020
|
|
6OX4
 
 | | A SETD3 Mutant (N255A) in Complex with an Actin Peptide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Actin Peptide, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2019-05-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
6OX3
 
 | | SETD3 in Complex with an Actin Peptide with His73 Replaced with Lysine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Actin Peptide, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2019-05-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.785 Å) | | Cite: | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
6OX1
 
 | | SETD3 in Complex with an Actin Peptide with Target Histidine Partially Methylated | | Descriptor: | 1,2-ETHANEDIOL, Actin, cytoplasmic 1, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2019-05-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
4Y4R
 
 | | Crystal structure of ribosomal oxygenase NO66 dimer mutant | | Descriptor: | ACETATE ION, Bifunctional lysine-specific demethylase and histidyl-hydroxylase NO66, NICKEL (II) ION | | Authors: | Wang, C, Hang, T, Zang, J. | | Deposit date: | 2015-02-11 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the JmjC domain-containing protein NO66 complexed with ribosomal protein Rpl8.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6OX5
 
 | | A SETD3 Mutant (N255A) in Complex with an Actin Peptide with His73 Replaced with Lysine | | Descriptor: | 1,2-ETHANEDIOL, Actin Peptide, Actin-histidine N-methyltransferase, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2019-05-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
6ZOV
 
 | | ENTEROPEPTIDASE IN COMPLEX WITH COMPOUND 6 | | Descriptor: | 1,2-ETHANEDIOL, 4-carbamimidamidobenzoic acid, Enteropeptidase, ... | | Authors: | Cummings, M.D. | | Deposit date: | 2020-07-07 | | Release date: | 2020-10-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Targeting Enteropeptidase with Reversible Covalent Inhibitors To Achieve Metabolic Benefits.
J.Pharmacol.Exp.Ther., 375, 2020
|
|
