6YQP
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual inhibitor TW22 | | Descriptor: | (~{E})-3-[4-[[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6 ),4,7,10,12-pentaen-9-yl]ethanoylamino]methyl]phenyl]-~{N}-oxidanyl-prop-2-enamide, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Joerger, A.C, Balourdas, D.I, Weiser, T, Chatterjee, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-17 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Characterization of a dual BET/HDAC inhibitor for treatment of pancreatic ductal adenocarcinoma.
Int.J.Cancer, 147, 2020
|
|
6YQO
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual inhibitor TW12 | | Descriptor: | (S)-N1-(4-(2-(4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl)acetamido)phenyl)-N8-hydroxyoctanediamide, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Joerger, A.C, Balourdas, D.I, Weiser, T, Chatterjee, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-17 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Characterization of a dual BET/HDAC inhibitor for treatment of pancreatic ductal adenocarcinoma.
Int.J.Cancer, 147, 2020
|
|
6T1L
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with piperazine-urea derivative 3 | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL, ~{N}-[[4-(diethylaminomethyl)phenyl]methyl]-4-pyrimidin-2-yl-piperazine-1-carboxamide | | Authors: | Chaikuad, A, Heidenreich, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
5BT4
 
 | | Crystal structure of BRD4 first bromodomain in complex with SGC-CBP30 chemical probe | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, Bromodomain-containing protein 4 | | Authors: | Tallant, C, Hay, D, Krojer, T, Nunez-Alonso, G, Picaud, S, Newman, J.A, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of BRD4 first bromodomain in complex with a 3,5-dimethylisoxazol ligand
To Be Published
|
|
5BT5
 
 | | Crystal structure of BRD2 second bromodomain in complex with SGC-CBP30 chemical probe | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, Bromodomain-containing protein 2 | | Authors: | Tallant, C, Hay, D, Krojer, T, Nunez-Alonso, G, Picaud, S, Newman, J.A, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of BRD2 second bromodomain in complex with a 3,5-dimethylisoxazol ligand
To Be Published
|
|
5C65
 
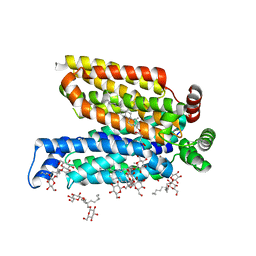 | | Structure of the human glucose transporter GLUT3 / SLC2A3 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Octyl Glucose Neopentyl Glycol, Solute carrier family 2, ... | | Authors: | Pike, A.C.W, Quigley, A, Chu, A, Tessitore, A, Xia, X, Mukhopadhyay, S, Wang, D, Kupinska, K, Strain-Damerell, C, Chalk, R, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-22 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the human glucose transporter GLUT3 / SLC2A3
To Be Published
|
|
5VT1
 
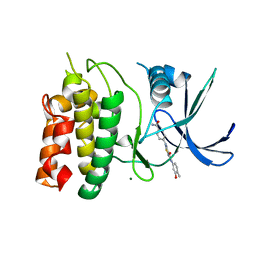 | | Crystal Structure of the Human CAMKK2B bound to a thiadiazinone benzamide inhibitor | | Descriptor: | 4-({5-[(3-hydroxy-4-methylphenyl)amino]-4-oxo-4H-1,2,6-thiadiazin-3-yl}amino)benzamide, Calcium/calmodulin-dependent protein kinase kinase 2, MAGNESIUM ION | | Authors: | Counago, R.M, Asquith, C.R.M, Arruda, P, Edwards, A.M, Gileadi, O, Kalogirou, A.S, Koutentis, P.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-05-15 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1,2,6-Thiadiazinones as Novel Narrow Spectrum Calcium/Calmodulin-Dependent Protein Kinase Kinase 2 (CaMKK2) Inhibitors.
Molecules, 23, 2018
|
|
5C8H
 
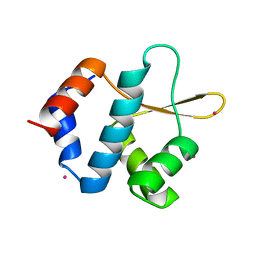 | | Crystal structure of ORC2 C-terminal domain | | Descriptor: | Origin recognition complex subunit 2, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Xu, C, Dong, A, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-25 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of ORC2 C-terminal domain
To Be Published
|
|
5WP3
 
 | | Crystal Structure of EED in complex with EB22 | | Descriptor: | EB22, Polycomb protein EED, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Zhu, L, Moody, J.D, Baker, D, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | First critical repressive H3K27me3 marks in embryonic stem cells identified using designed protein inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6OYY
 
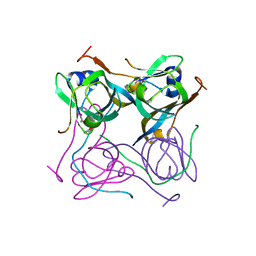 | | Crystal structure of Mtb aspartate decarboxylase, pyrazinoic acid complex | | Descriptor: | Aspartate 1-decarboxylase alpha chain, Aspartate 1-decarboxylase beta chain, PYRAZINE-2-CARBOXYLIC ACID | | Authors: | Sun, Q, Li, X, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2019-05-15 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The molecular basis of pyrazinamide activity on Mycobacterium tuberculosis PanD.
Nat Commun, 11, 2020
|
|
6BHG
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | Descriptor: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-10-30 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
6P02
 
 | | Crystal structure of Mtb aspartate decarboxylase, 6-Chlorine pyrazinoic acid complex | | Descriptor: | 6-chloropyrazine-2-carboxylic acid, Aspartate 1-decarboxylase alpha chain, Aspartate 1-decarboxylase beta chain | | Authors: | Sun, Q, Li, X, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2019-05-16 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The molecular basis of pyrazinamide activity on Mycobacterium tuberculosis PanD.
Nat Commun, 11, 2020
|
|
6BHD
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | Descriptor: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, SODIUM ION, ... | | Authors: | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-10-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
5WOF
 
 | | 1.65 ANGSTROM STRUCTURE OF THE DYNEIN LIGHT CHAIN 1 FROM PLASMODIUM FALCIPARUM | | Descriptor: | Dynein light chain 1, putative | | Authors: | Walker, J.R, Lew, J, Amani, M, Alam, Z, Wasney, G, Boulanger, K, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Hui, R, Botchkarev, A, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Genome-scale Protein Expression and Structural Biology of
Plasmodium Falciparum and Related Apicomplexan Organisms.
MOL.BIOCHEM.PARASITOL., 151, 2007
|
|
6OZ8
 
 | |
6PED
 
 | | Crystal structure of HEMK2-TRMT112 complex | | Descriptor: | Methyltransferase N6AMT1, Multifunctional methyltransferase subunit TRM112-like protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-20 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of HEMK2-TRMT112 complex
To Be Published
|
|
3BEY
 
 | | Crystal structure of the protein O27018 from Methanobacterium thermoautotrophicum. Northeast Structural Genomics Consortium target TT217 | | Descriptor: | Conserved protein O27018 | | Authors: | Kuzin, A.P, Gu, J, Xu, X, Neely, H, Forouhar, F, Owens, L, Mao, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-20 | | Release date: | 2007-12-18 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the protein O27018 from Methanobacterium thermoautotrophicum.
To be Published
|
|
8QWO
 
 | | Structure of p53 cancer mutant Y234C | | Descriptor: | Cellular tumor antigen p53, GLYCEROL, ZINC ION | | Authors: | Balourdas, D.I, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
8QWM
 
 | | Structure of p53 cancer mutant Y205C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, L(+)-TARTARIC ACID, ... | | Authors: | Balourdas, D.I, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
8QWN
 
 | | Structure of p53 cancer mutant Y220C (hexagonal crystal form) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cellular tumor antigen p53, ... | | Authors: | Balourdas, D.I, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
8QWL
 
 | | Structure of p53 cancer mutant Y163C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, MALONATE ION, ... | | Authors: | Balourdas, D.I, Markl, A.M, Kraemer, A, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
6PDM
 
 | | Crystal structure of Human Protein Arginine Methyltransferase 9 (PRMT9) | | Descriptor: | Protein arginine N-methyltransferase 9, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Tempel, W, Zeng, H, Li, Y, Seitova, A, Hutchinson, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-19 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Human Protein Arginine Methyltransferase 9 (PRMT9)
To Be Published
|
|
8QWK
 
 | | Structure of p53 cancer mutant Y126C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, ZINC ION | | Authors: | Markl, A.M, Balourdas, D.I, Kraemer, A, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
8QWP
 
 | | Structure of p53 cancer mutant Y236C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, L(+)-TARTARIC ACID, ... | | Authors: | Balourdas, D.I, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
3BID
 
 | | Crystal structure of the NMB1088 protein from Neisseria meningitidis. Northeast Structural Genomics Consortium target MR91 | | Descriptor: | UPF0339 protein NMB1088 | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Mao, L, Fang, Y, Xiao, R, Owen, L.A, Maglaqui, M, Cunningham, K, Baran, M.C, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-18 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the NMB1088 protein from Neisseria meningitidis.
To be Published
|
|
