3H0I
 
 | |
1H1Q
 
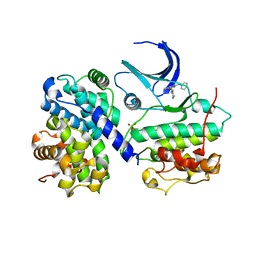 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with the inhibitor NU6094 | | Descriptor: | 2-ANILINO-6-CYCLOHEXYLMETHOXYPURINE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Davies, T.G, Noble, M.E.M, Endicott, J.A, Johnson, L.N. | | Deposit date: | 2002-07-21 | | Release date: | 2002-09-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of a Potent Purine-Based Cyclin-Dependent Kinase Inhibitor
Nat.Struct.Biol., 9, 2002
|
|
4KFN
 
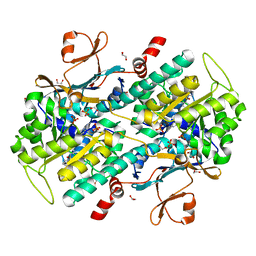 | | Structure-Based Discovery of Novel Amide-Containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-[4-(piperidin-1-ylsulfonyl)benzyl]-1H-pyrrolo[3,2-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynolds, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Gunzner-Toste, J, Liederer, B.M, Ly, J, O'Brien, T, Oh, A, Wang, L, Wang, W, Xiao, Y, Zak, M, Zhao, G, Yuen, P, Bair, K.W. | | Deposit date: | 2013-04-27 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based identification of ureas as novel nicotinamide phosphoribosyltransferase (nampt) inhibitors.
J.Med.Chem., 56, 2013
|
|
1H1S
 
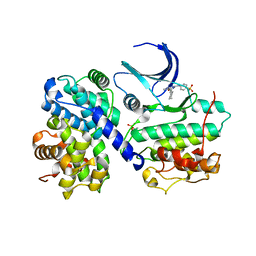 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with the inhibitor NU6102 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, O6-CYCLOHEXYLMETHOXY-2-(4'-SULPHAMOYLANILINO) PURINE | | Authors: | Davies, T.G, Noble, M.E.M, Endicott, J.A, Johnson, L.N. | | Deposit date: | 2002-07-21 | | Release date: | 2002-09-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of a Potent Purine-Based Cyclin-Dependent Kinase Inhibitor
Nat.Struct.Biol., 9, 2002
|
|
1H1P
 
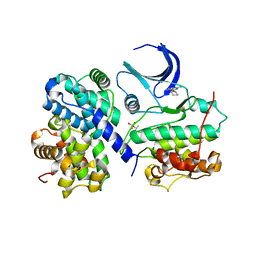 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with the inhibitor NU2058 | | Descriptor: | 6-O-CYCLOHEXYLMETHYL GUANINE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Davies, T.G, Noble, M.E.M, Endicott, J.A, Johnson, L.N. | | Deposit date: | 2002-07-21 | | Release date: | 2002-09-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of a Potent Purine-Based Cyclin-Dependent Kinase Inhibitor
Nat.Struct.Biol., 9, 2002
|
|
1H1R
 
 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with the inhibitor NU6086 | | Descriptor: | 6-CYCLOHEXYLMETHOXY-2-(3'-CHLOROANILINO) PURINE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Davies, T.G, Noble, M.E.M, Endicott, J.A, Johnson, L.N. | | Deposit date: | 2002-07-21 | | Release date: | 2002-09-19 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of a potent purine-based cyclin-dependent kinase inhibitor.
Nat. Struct. Biol., 9, 2002
|
|
2RL5
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a 2,3-dihydro-1,4-benzoxazine inhibitor | | Descriptor: | N-(4-CHLOROPHENYL)-7-[(6,7-DIMETHOXYQUINOLIN-4-YL)OXY]-2,3-DIHYDRO-1,4-BENZOXAZINE-4-CARBOXAMIDE, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Long, A.M, Rose, P, Zhao, H. | | Deposit date: | 2007-10-18 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Novel 2,3-dihydro-1,4-benzoxazines as potent and orally bioavailable inhibitors of tumor-driven angiogenesis.
J.Med.Chem., 51, 2008
|
|
4J6I
 
 | | Discovery of thiazolobenzoxepin PI3-kinase inhibitors that spare the PI3-kinase beta isoform | | Descriptor: | 2-methyl-1-(4-{2-[1-(2,2,2-trifluoroethyl)-1H-1,2,4-triazol-5-yl]-4,5-dihydro[1]benzoxepino[5,4-d][1,3]thiazol-8-yl}-1H-pyrazol-1-yl)propan-2-ol, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Rouge, L, Wu, P. | | Deposit date: | 2013-02-11 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of thiazolobenzoxepin PI3-kinase inhibitors that spare the PI3-kinase beta isoform.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7CWN
 
 | | P17-H014 Fab cocktail in complex with SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, N, Wang, X. | | Deposit date: | 2020-08-29 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection.
Cell Res., 31, 2021
|
|
8YFN
 
 | |
8YFM
 
 | | Crystal structure of FIP200 claw/TNIP1_FIR_pS122 | | Descriptor: | GLYCEROL, RB1-inducible coiled-coil protein 1, SULFATE ION, ... | | Authors: | Lv, M.Q, Wu, S.M. | | Deposit date: | 2024-02-24 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for TNIP1 binding to FIP200 during mitophagy.
J.Biol.Chem., 300, 2024
|
|
8YFL
 
 | | crystal structure of FIP200 claw/TNIP1_FIR_pS122pS123 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, RB1-inducible coiled-coil protein 1, ... | | Authors: | Lv, M.Q, Wu, S.M. | | Deposit date: | 2024-02-24 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for TNIP1 binding to FIP200 during mitophagy.
J.Biol.Chem., 300, 2024
|
|
8YFK
 
 | | Crystal structure of FIP200 claw/TNIP1_FIR_pS123 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, RB1-inducible coiled-coil protein 1, ... | | Authors: | Lv, M.Q, Wu, S.M. | | Deposit date: | 2024-02-24 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for TNIP1 binding to FIP200 during mitophagy.
J.Biol.Chem., 300, 2024
|
|
8XJV
 
 | | Structural basis for the linker histone H5-nucleosome binding and chromatin compaction | | Descriptor: | DNA, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, W.Y, Song, F, Zhu, P. | | Deposit date: | 2023-12-22 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for linker histone H5-nucleosome binding and chromatin fiber compaction.
Cell Res., 34, 2024
|
|
4OCH
 
 | | Apo structure of Smr domain of MutS2 from Deinococcus radiodurans | | Descriptor: | Endonuclease MutS2, GLYCEROL | | Authors: | Zhang, H, Zhao, Y, Xu, Q, Hua, Y.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4001 Å) | | Cite: | Structural and functional studies of MutS2 from Deinococcus radiodurans.
Dna Repair, 21, 2014
|
|
4OD6
 
 | | Structure of Smr domain of MutS2 from Deinococcus radiodurans, Mn2+ soaked | | Descriptor: | Endonuclease MutS2 | | Authors: | Zhang, H, Zhao, Y, Xu, Q, Hua, Y.J. | | Deposit date: | 2014-01-10 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Structural and functional studies of MutS2 from Deinococcus radiodurans.
Dna Repair, 21, 2014
|
|
2AX5
 
 | | Solution Structure of Urm1 from Saccharomyces Cerevisiae | | Descriptor: | Hypothetical 11.0 kDa protein in FAA3-MAS3 intergenic region | | Authors: | Xu, J, Huang, H, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2005-09-03 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Urm1 and its implications for the origin of protein modifiers.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7XAZ
 
 | | Crystal structure of Omicron BA.1.1 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liao, H, Meng, Y, Li, W. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
7XB1
 
 | | Crystal structure of Omicron BA.3 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, W, Meng, Y, Liao, H. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
7CWO
 
 | | SARS-CoV-2 spike protein RBD and P17 fab complex | | Descriptor: | Spike glycoprotein, heavy chain of P17 Fab, light chain of P17 Fab | | Authors: | Wang, X, Wang, N. | | Deposit date: | 2020-08-29 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection.
Cell Res., 31, 2021
|
|
7CMW
 
 | | Complex structure of PARP1 catalytic domain with pamiparib | | Descriptor: | (2R)-14-fluoro-2-methyl-6,9,10,19-tetrazapentacyclo[14.2.1.02,6.08,18.012,17]nonadeca-1(18),8,12(17),13,15-pentaen-11-one, GLYCEROL, Poly [ADP-ribose] polymerase 1 | | Authors: | Feng, Y.C, Peng, H, Hong, Y, Liu, Y. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Pamiparib (BGB-290), a Potent and Selective Poly (ADP-ribose) Polymerase (PARP) Inhibitor in Clinical Development.
J.Med.Chem., 63, 2020
|
|
7CWL
 
 | |
7CWM
 
 | |
7CWU
 
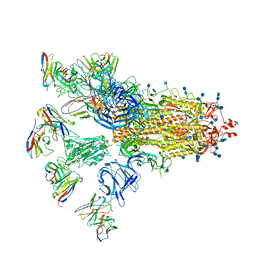 | | SARS-CoV-2 spike proteins trimer in complex with P17 and FC05 Fabs cocktail | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Wang, N. | | Deposit date: | 2020-08-31 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure-based development of human antibody cocktails against SARS-CoV-2.
Cell Res., 31, 2021
|
|
2EWP
 
 | |
