6TY9
 
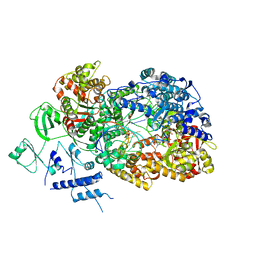 | | In situ structure of BmCPV RNA dependent RNA polymerase at initiation state | | Descriptor: | MAGNESIUM ION, Non-template RNA (5'-D(*(GTA))-R(P*GP*UP*AP*AP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), RNA-dependent RNA Polymerase, ... | | Authors: | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2019-08-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6TZ1
 
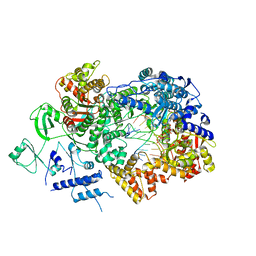 | | In situ structure of BmCPV RNA-dependent RNA polymerase at early-elongation state | | Descriptor: | Non-template RNA (5'-D(*(GTA))-R(P*GP*UP*A)-3'), RNA-dependent RNA Polymerase, Template RNA (5'-R(P*AP*GP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2019-08-09 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6TY8
 
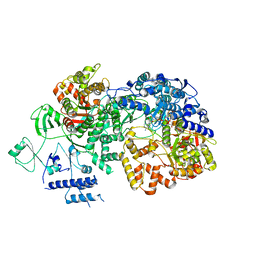 | | In situ structure of BmCPV RNA dependent RNA polymerase at quiescent state | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA-dependent RNA Polymerase, Viral structural protein 4 | | Authors: | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2019-08-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6TZ2
 
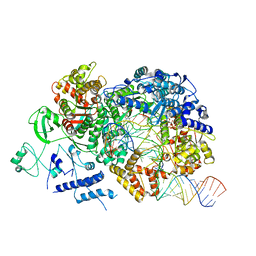 | | In situ structure of BmCPV RNA-dependent RNA polymerase at elongation state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Non-template RNA (36-MER), ... | | Authors: | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2019-08-09 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6TZ0
 
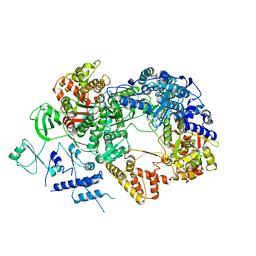 | | In situ structure of BmCPV RNA-dependent RNA polymerase at abortive state | | Descriptor: | RNA-dependent RNA Polymerase, Viral structural protein 4 | | Authors: | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2019-08-09 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6UX2
 
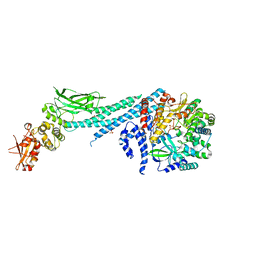 | | Crystal structure of ZIKV RdRp in complex with STAT2 | | Descriptor: | Nonstructural Protein 5, SULFATE ION, Signal transducer and activator of transcription 2, ... | | Authors: | Wang, B, Song, J. | | Deposit date: | 2019-11-06 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WCZ
 
 | | CryoEM structure of full-length ZIKV NS5-hSTAT2 complex | | Descriptor: | Non-structural protein 5, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Boxiao, W, Stephanie, T, Kang, Z, Maria, T.S, Jian, F, Jiuwei, L, Linfeng, G, Wendan, R, Yanxiang, C, Ethan, C.V, HeaJin, H, Matthew, J.E, Sean, E.O, Adolfo, G.S, Hong, Z, Rong, H, Jikui, S. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1HR2
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A MUTANT P4-P6 DOMAIN (DELC209) OF TETRAHYMENA THEMOPHILA GROUP I INTRON. | | Descriptor: | MAGNESIUM ION, P4-P6 DELC209 MUTANT RNA RIBOZYME DOMAIN | | Authors: | Juneau, K, Podell, E.R, Harrington, D.J, Cech, T.R. | | Deposit date: | 2000-12-20 | | Release date: | 2001-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA--solvent interactions.
Structure, 9, 2001
|
|
4N0S
 
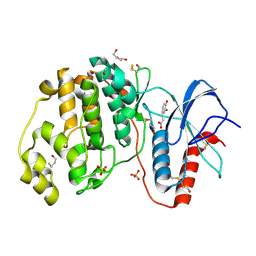 | | Complex of ERK2 with caffeic acid | | Descriptor: | CAFFEIC ACID, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Kurinov, I, Malakhova, M. | | Deposit date: | 2013-10-02 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7992 Å) | | Cite: | Caffeic Acid Directly Targets ERK1/2 to Attenuate Solar UV-Induced Skin Carcinogenesis.
Cancer Prev Res (Phila), 7, 2014
|
|
376D
 
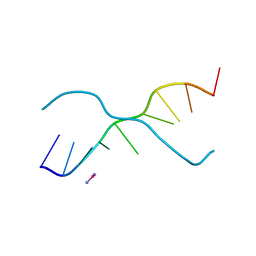 | | A ZIPPER-LIKE DNA DUPLEX D(GCGAAAGCT) | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*(CBR)P*GP*AP*AP*AP*GP*CP*T)-3') | | Authors: | Cruse, W.B.T, Shepard, W, Prange, T, delalFortelle, E, Fourme, R. | | Deposit date: | 1998-01-22 | | Release date: | 1999-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A zipper-like duplex in DNA: the crystal structure of d(GCGAAAGCT) at 2.1 A resolution.
Structure, 6, 1998
|
|
6LH8
 
 | | Structure of aerolysin-like protein (Bombina maxima) | | Descriptor: | aerolysin-like protein | | Authors: | Bian, X.L, Wang, Q.Q, Li, X, Teng, M.Q, Zhang, Y. | | Deposit date: | 2019-12-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | A cellular endolysosome-modulating pore-forming protein from a toad is negatively regulated by its paralog under oxidizing conditions.
J.Biol.Chem., 295, 2020
|
|
6LHZ
 
 | | Structure of aerolysin-like protein (Bombina maxima) | | Descriptor: | aerolysin-like protein | | Authors: | Bian, X.L, Wang, Q.Q, Li, X, Teng, M.Q, Zhang, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | A cellular endolysosome-modulating pore-forming protein from a toad is negatively regulated by its paralog under oxidizing conditions.
J.Biol.Chem., 295, 2020
|
|
5D2S
 
 | |
5D2Q
 
 | |
2FFL
 
 | |
5D23
 
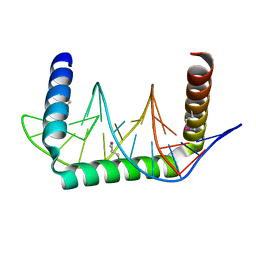 | |
8XGC
 
 | | Structure of yeast replisome associated with FACT and histone hexamer, Composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, Chromosome segregation in meiosis protein 3, ... | | Authors: | Li, N, Gao, Y, Yu, D, Gao, N, Zhai, Y. | | Deposit date: | 2023-12-15 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Parental histone transfer caught at the replication fork.
Nature, 627, 2024
|
|
6OBH
 
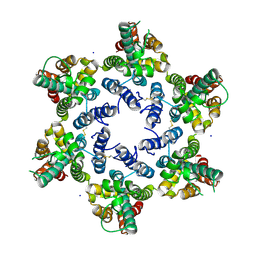 | | Structure of HIV-1 CA 1/2-hexamer | | Descriptor: | CA, SODIUM ION | | Authors: | Summers, B.J, Xiong, Y. | | Deposit date: | 2019-03-20 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
1GRZ
 
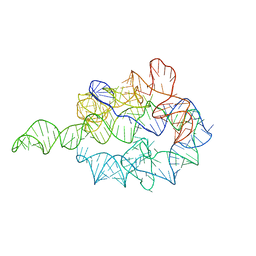 | |
5HMP
 
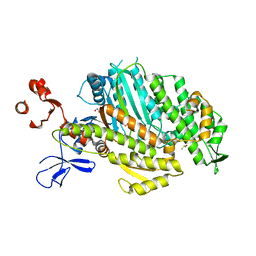 | | Myosin Vc pre-powerstroke state | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Ropars, V, Pylypenko, O, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2016-01-16 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | The myosin X motor is optimized for movement on actin bundles.
Nat Commun, 7, 2016
|
|
5I0I
 
 | | Crystal structure of myosin X motor domain with 2IQ motifs in pre-powerstroke state | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, Calmodulin, ... | | Authors: | Isabet, T, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2016-02-04 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The myosin X motor is optimized for movement on actin bundles.
Nat Commun, 7, 2016
|
|
5I0H
 
 | | Crystal structure of myosin X motor domain in pre-powerstroke state | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Isabet, T, Blanc, F, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2016-02-04 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The myosin X motor is optimized for movement on actin bundles.
Nat Commun, 7, 2016
|
|
5HMO
 
 | |
5KG8
 
 | |
6UFR
 
 | | Structure of recombinantly assembled E46K alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein | | Authors: | Eisenberg, D.S, Boyer, D.R, Sawaya, M.R, Li, B, Jiang, L. | | Deposit date: | 2019-09-24 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The alpha-synuclein hereditary mutation E46K unlocks a more stable, pathogenic fibril structure.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
