1Q6A
 
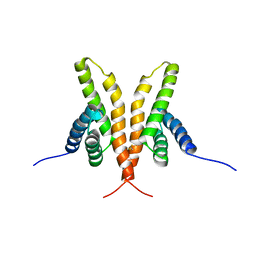 | | Solution Structure of the C-terminal Domain of Thermosynechococcus elongatus KaiA (ThKaiA180C); Averaged Minimized Structure | | Descriptor: | Circadian clock protein KaiA homolog | | Authors: | Vakonakis, I, Sun, J, Holzenburg, A, Golden, S.S, LiWang, A.C. | | Deposit date: | 2003-08-13 | | Release date: | 2003-08-19 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the KaiC-interacting C-terminal domain of KaiA, a circadian clock protein: Implications for KaiA-KaiC interaction
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
8S9M
 
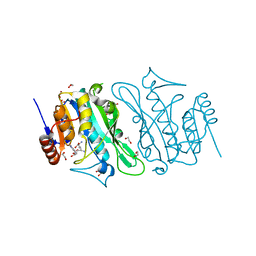 | | DNA cytosine-N4 methyltransferase (residues 79-324) from the Bdelloid rotifer Adineta vaga | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
8S9N
 
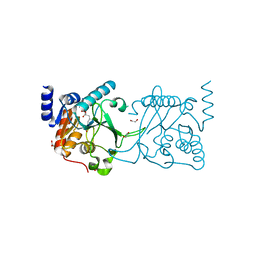 | | DNA cytosine-N4 methyltransferase (residues 61-324) from the Bdelloid rotifer Adineta vaga - C2 crystal form | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
8S9O
 
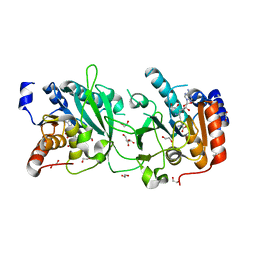 | | DNA cytosine-N4 methyltransferase (residues 61-324) from the Bdelloid rotifer Adineta vaga - P1 crystal form | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
6MT0
 
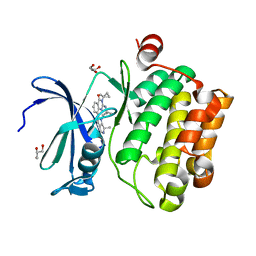 | | Crystal structure of human Pim-1 kinase in complex with a quinazolinone-pyrrolodihydropyrrolone inhibitor | | Descriptor: | 3-(1-methylcyclopropyl)-2-[(1-methylcyclopropyl)amino]-8-[(6R)-6-methyl-4-oxo-1,4,5,6-tetrahydropyrrolo[3,4-b]pyrrol-2-yl]quinazolin-4(3H)-one, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Mohr, C. | | Deposit date: | 2018-10-18 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of ( R)-8-(6-Methyl-4-oxo-1,4,5,6-tetrahydropyrrolo[3,4- b]pyrrol-2-yl)-3-(1-methylcyclopropyl)-2-((1-methylcyclopropyl)amino)quinazolin-4(3 H)-one, a Potent and Selective Pim-1/2 Kinase Inhibitor for Hematological Malignancies.
J. Med. Chem., 62, 2019
|
|
4DK5
 
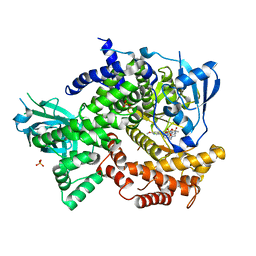 | | Crystal structure of human PI3K-gamma in complex with a pyridyl-triazine inhibitor | | Descriptor: | 4-(2-[(6-methoxypyridin-3-yl)amino]-5-{[4-(methylsulfonyl)piperazin-1-yl]methyl}pyridin-3-yl)-6-methyl-1,3,5-triazin-2-amine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2012-02-03 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-based design of a novel series of potent, selective inhibitors of the class I phosphatidylinositol 3-kinases.
J.Med.Chem., 55, 2012
|
|
5X6O
 
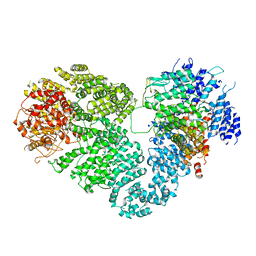 | | Intact ATR/Mec1-ATRIP/Ddc2 complex | | Descriptor: | DNA damage checkpoint protein LCD1, Serine/threonine-protein kinase MEC1 | | Authors: | Wang, X, Ran, T, Cai, G. | | Deposit date: | 2017-02-22 | | Release date: | 2017-12-20 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | 3.9 angstrom structure of the yeast Mec1-Ddc2 complex, a homolog of human ATR-ATRIP.
Science, 358, 2017
|
|
6LQA
 
 | | voltage-gated sodium channel Nav1.5 with quinidine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Quinidine, Sodium channel protein type 5 subunit alpha | | Authors: | Yan, N, Li, Z, Pan, X, Huang, G. | | Deposit date: | 2020-01-13 | | Release date: | 2021-03-24 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Basis for Pore Blockade of the Human Cardiac Sodium Channel Na v 1.5 by the Antiarrhythmic Drug Quinidine*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
3LRU
 
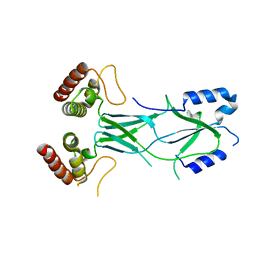 | |
8FHS
 
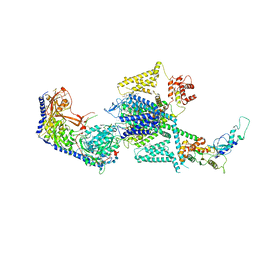 | | Human L-type voltage-gated calcium channel Cav1.2 in the presence of amiodarone and sofosbuvir at 3.3 Angstrom resolution | | Descriptor: | (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
6B9M
 
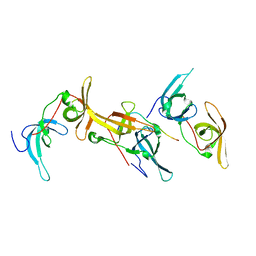 | |
4UMT
 
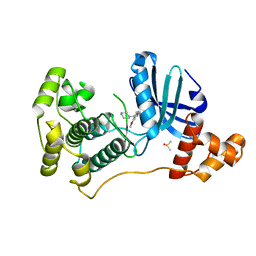 | | Structure of MELK in complex with inhibitors | | Descriptor: | 1-(4-{[3-(isoquinolin-7-yl)prop-2-yn-1-yl]oxy}-2-methoxybenzyl)piperazinediium, DIMETHYL SULFOXIDE, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-21 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-Based Design of Type II Inhibitors Applied to Maternal Embryonic Leucine Zipper Kinase.
Acs Med.Chem.Lett., 6, 2015
|
|
4UMU
 
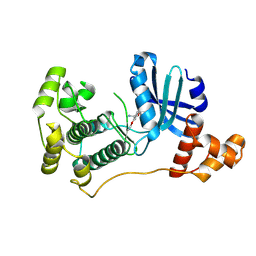 | | Structure of MELK in complex with inhibitors | | Descriptor: | (2-ethoxy-4-{[3-(isoquinolin-7-yl)prop-2-yn-1-yl]oxy}phenyl)methanaminium, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-21 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-Based Design of Type II Inhibitors Applied to Maternal Embryonic Leucine Zipper Kinase.
Acs Med.Chem.Lett., 6, 2015
|
|
6LGN
 
 | | The atomic structure of varicella zoster virus C-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Li, S, Zheng, Q. | | Deposit date: | 2019-12-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
6LGL
 
 | | The atomic structure of varicella-zoster virus A-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Zheng, Q, Li, S. | | Deposit date: | 2019-12-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
7F17
 
 | | Crystal Structure of acid phosphatase | | Descriptor: | Acid phosphatase | | Authors: | Xu, X, Hou, X.D, Song, W, Rao, Y.J, Liu, L.M, Wu, J. | | Deposit date: | 2021-06-08 | | Release date: | 2021-10-27 | | Last modified: | 2022-05-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Local Electric Field Modulated Reactivity of Pseudomonas aeruginosa Acid Phosphatase for Enhancing Phosphorylation of l-Ascorbic Acid
Acs Catalysis, 11, 2021
|
|
7XH6
 
 | | Crystal structure of CBP bromodomain liganded with CCS1477 | | Descriptor: | (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, CREB-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-04-07 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
7XHE
 
 | | Crystal structure of CBP bromodomain liganded with CCS151 | | Descriptor: | (6S)-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(3R)-1-methylsulfonylpyrrolidin-3-yl]benzimidazol-2-yl]-1-(3-fluoranyl-4-methoxy-phenyl)piperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein | | Authors: | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-04-08 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
7XI0
 
 | | Crystal structure of CBP bromodomain liganded with CCS150 | | Descriptor: | (6S)-1-(3-chloranyl-4-methoxy-phenyl)-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(3R)-1-methylsulfonylpyrrolidin-3-yl]benzimidazol-2-yl]piperidin-2-one, CREB-binding protein, GLYCEROL | | Authors: | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-04-11 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
7XVF
 
 | | Nav1.7 mutant class2 | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Huang, G, Wu, Q, Li, Z, Pan, X, Yan, N. | | Deposit date: | 2022-05-22 | | Release date: | 2022-08-10 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Unwinding and spiral sliding of S4 and domain rotation of VSD during the electromechanical coupling in Na v 1.7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XVE
 
 | | Human Nav1.7 mutant class-I | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Huang, G, Wu, Q, Li, Z, Pan, X, Yan, N. | | Deposit date: | 2022-05-21 | | Release date: | 2022-08-10 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Unwinding and spiral sliding of S4 and domain rotation of VSD during the electromechanical coupling in Na v 1.7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2NBQ
 
 | |
2PL9
 
 | |
2PMC
 
 | |
7F18
 
 | | Crystal Structure of a mutant of acid phosphatase from Pseudomonas aeruginosa (Q57H/W58P/D135R) | | Descriptor: | Acid phosphatase | | Authors: | Xu, X, Hou, X.D, Song, W, Yin, D.J, Rao, Y.J, Liu, L.M. | | Deposit date: | 2021-06-08 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Local Electric Field Modulated Reactivity of Pseudomonas aeruginosa Acid Phosphatase for Enhancing Phosphorylation of l-Ascorbic Acid
Acs Catalysis, 11, 2021
|
|
