6RQW
 
 | |
6RQN
 
 | |
6RQU
 
 | |
6RQQ
 
 | |
8FFI
 
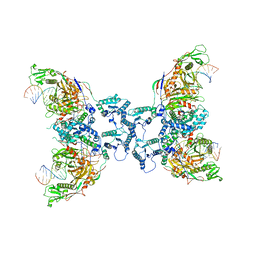 | | Structure of tetramerized MapSPARTA upon guide RNA-mediated target DNA binding | | Descriptor: | MAGNESIUM ION, TIR-APAZ, guide RNA, ... | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2022-12-08 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Oligomerization-mediated activation of a short prokaryotic Argonaute.
Nature, 621, 2023
|
|
8Y65
 
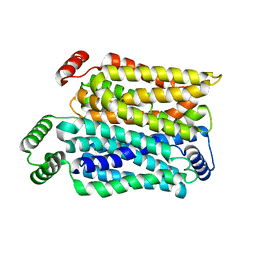 | | Cryo-EM structure of human urate transporter GLUT9 bound to substrate urate | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 9, URIC ACID | | Authors: | Pan, X.J, Shen, Z.L, Xu, L, Huang, G.X.Y. | | Deposit date: | 2024-02-01 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural basis for urate recognition and apigenin inhibition of human GLUT9.
Nat Commun, 15, 2024
|
|
8Y66
 
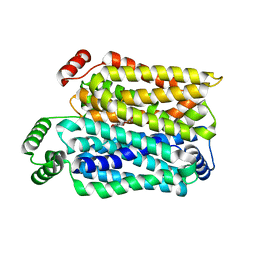 | | Cryo-EM structure of human urate transporter GLUT9 bound to inhibitor apigenin | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Solute carrier family 2, facilitated glucose transporter member 9 | | Authors: | Pan, X.J, Shen, Z.L, Xu, L, Huang, G.X.Y. | | Deposit date: | 2024-02-01 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for urate recognition and apigenin inhibition of human GLUT9.
Nat Commun, 15, 2024
|
|
7LBR
 
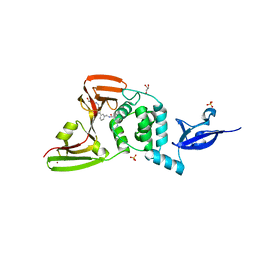 | | SARS-CoV-2 papain-like protease (PLpro) bound to inhibitor XR8-89 | | Descriptor: | 5-[(azetidin-3-yl)amino]-N-[(1R)-1-{3-[5-({[(1S,3R)-3-hydroxycyclopentyl]amino}methyl)thiophen-2-yl]phenyl}ethyl]-2-methylbenzamide, GLYCEROL, Non-structural protein 3, ... | | Authors: | Ratia, K.M, Xiong, R, Thatcher, G.R. | | Deposit date: | 2021-01-08 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Potent, Novel SARS-CoV-2 PLpro Inhibitors Block Viral Replication in Monkey and Human Cell Cultures.
Biorxiv, 2021
|
|
7LBS
 
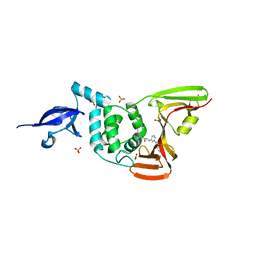 | | SARS-CoV-2 papain-like protease (PLpro) bound to inhibitor XR8-24 | | Descriptor: | 5-[(azetidin-3-yl)amino]-2-methyl-N-[(1R)-1-(3-{5-[(pyrrolidin-1-yl)methyl]thiophen-2-yl}phenyl)ethyl]benzamide, BORIC ACID, GLYCEROL, ... | | Authors: | Ratia, K.M, Xiong, R, Thatcher, G.R. | | Deposit date: | 2021-01-08 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Potent, Novel SARS-CoV-2 PLpro Inhibitors Block Viral Replication in Monkey and Human Cell Cultures.
Biorxiv, 2021
|
|
7LLF
 
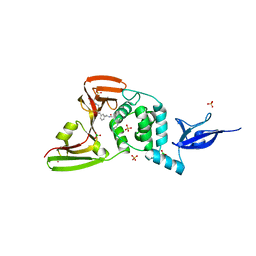 | | SARS-CoV-2 papain-like protease (PLpro) bound to inhibitor XR8-83 | | Descriptor: | 5-[(azetidin-3-yl)amino]-N-[(1R)-1-{3-[5-({[(1R,3S)-3-hydroxycyclopentyl]amino}methyl)thiophen-2-yl]phenyl}ethyl]-2-methylbenzamide, BORIC ACID, GLYCEROL, ... | | Authors: | Ratia, K.M, Xiong, R, Thatcher, G.R. | | Deposit date: | 2021-02-03 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent, Novel SARS-CoV-2 PLpro Inhibitors Block Viral Replication in Monkey and Human Cell Cultures.
Biorxiv, 2021
|
|
7LOS
 
 | | SARS-CoV-2 papain-like protease (PLpro) bound to inhibitor XR8-65 | | Descriptor: | 5-(azetidin-3-ylamino)-2-methyl-~{N}-[(1~{R})-1-[3-[5-[[[(3~{R})-oxolan-3-yl]amino]methyl]thiophen-2-yl]phenyl]ethyl]benzamide, CHLORIDE ION, Non-structural protein 3, ... | | Authors: | Ratia, K.M, Xiong, R, Thatcher, G.R. | | Deposit date: | 2021-02-10 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Potent, Novel SARS-CoV-2 PLpro Inhibitors Block Viral Replication in Monkey and Human Cell Cultures.
Biorxiv, 2021
|
|
7LLZ
 
 | | SARS-CoV-2 papain-like protease (PLpro) bound to inhibitor XR8-69 | | Descriptor: | N-[(1R)-1-(3-{5-[(acetylamino)methyl]thiophen-2-yl}phenyl)ethyl]-5-[(azetidin-3-yl)amino]-2-methylbenzamide, Non-structural protein 3, SULFATE ION, ... | | Authors: | Ratia, K.M, Xiong, R, Thatcher, G.R. | | Deposit date: | 2021-02-04 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Potent, Novel SARS-CoV-2 PLpro Inhibitors Block Viral Replication in Monkey and Human Cell Cultures.
Biorxiv, 2021
|
|
2LJ7
 
 | |
2LG4
 
 | |
3DEZ
 
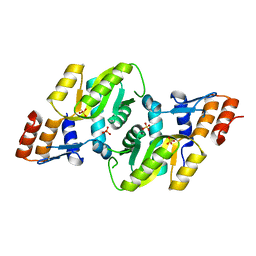 | | Crystal structure of Orotate phosphoribosyltransferase from Streptococcus mutans | | Descriptor: | Orotate phosphoribosyltransferase, SULFATE ION | | Authors: | Liu, C.P, Gao, Z.Q, Hou, H.F, Li, L.F, Su, X.D, Dong, Y.H. | | Deposit date: | 2008-06-11 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of orotate phosphoribosyltransferase from the caries pathogen Streptococcus mutans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
4HZ9
 
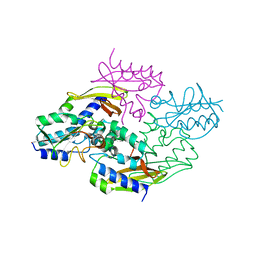 | | Crystal structure of the type VI native effector-immunity complex Tae3-Tai3 from Ralstonia pickettii | | Descriptor: | Putative cytoplasmic protein, Putative periplasmic protein | | Authors: | Dong, C, Zhang, H, Gao, Z.Q, Dong, Y.H. | | Deposit date: | 2012-11-14 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the inhibition of type VI effector Tae3 by its immunity protein Tai3
Biochem.J., 454, 2013
|
|
4HZB
 
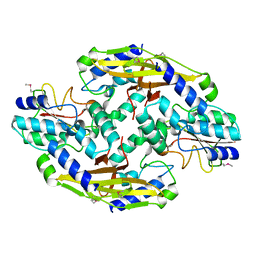 | | Crystal structure of the type VI SeMet effector-immunity complex Tae3-Tai3 from Ralstonia pickettii | | Descriptor: | Putative cytoplasmic protein, Putative periplasmic protein | | Authors: | Dong, C, Zhang, H, Gao, Z.Q, Dong, Y.H. | | Deposit date: | 2012-11-15 | | Release date: | 2013-08-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the inhibition of type VI effector Tae3 by its immunity protein Tai3
Biochem.J., 454, 2013
|
|
8HDL
 
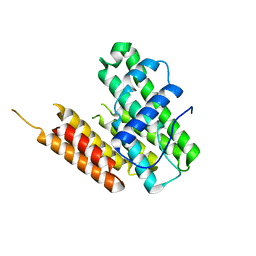 | |
7CHU
 
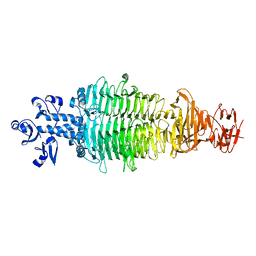 | | Geobacillus virus E2 - ORF18 | | Descriptor: | Putative pectin lyase | | Authors: | Gong, Y. | | Deposit date: | 2020-07-06 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | Structural and functional characterization of the deep-sea thermophilic bacteriophage GVE2 tailspike protein.
Int.J.Biol.Macromol., 164, 2020
|
|
7V55
 
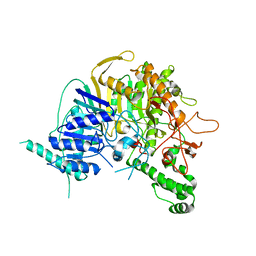 | |
7V53
 
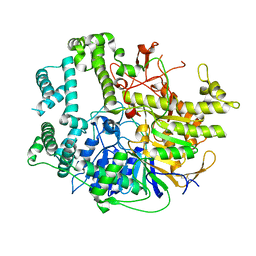 | |
7WDK
 
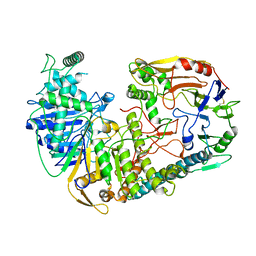 | | The structure of PldA-PA3488 complex | | Descriptor: | Phospholipase D, Tli4_C domain-containing protein | | Authors: | Zhao, L, Yang, X.Y, Li, Z.Q. | | Deposit date: | 2021-12-21 | | Release date: | 2022-10-26 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insights into PA3488-mediated inactivation of Pseudomonas aeruginosa PldA
Nat Commun, 13, 2022
|
|
7RW2
 
 | |
6ZCT
 
 | | Nonstructural protein 10 (nsp10) from SARS CoV-2 | | Descriptor: | ZINC ION, nsp10 | | Authors: | Rogstam, A, Nyblom, M, Christensen, S, Sele, C, Lindvall, T, Rasmussen, A.A, Andre, I, Fisher, S.Z, Knecht, W, Kozielski, F. | | Deposit date: | 2020-06-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of Non-Structural Protein 10 from Severe Acute Respiratory Syndrome Coronavirus-2.
Int J Mol Sci, 21, 2020
|
|
9BF1
 
 | | Structure of apo-state V. cholerae DdmD | | Descriptor: | Helicase/UvrB N-terminal domain-containing protein | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2024-04-16 | | Release date: | 2024-08-21 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | DdmDE eliminates plasmid invasion by DNA-guided DNA targeting.
Cell, 187, 2024
|
|
