2K8O
 
 | |
6VEJ
 
 | | TriABC transporter from Pseudomonas aeruginosa | | Descriptor: | (2R)-2-(hexadecanoyloxy)propyl nonadecanoate, DODECYL-ALPHA-D-MALTOSIDE, PALMITIC ACID, ... | | Authors: | Sygusch, J, Fabre, L, Rouiller, I, Bhattacharyya, S. | | Deposit date: | 2020-01-02 | | Release date: | 2020-09-30 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | A "Drug Sweeping" State of the TriABC Triclosan Efflux Pump from Pseudomonas aeruginosa.
Structure, 29, 2021
|
|
5ZY8
 
 | | Crystal structure of C terminal truncated HadBC (3R-Hydroxyacyl-ACP Dehydratase) complex from Mycobacterium tuberculosis | | Descriptor: | 3-hydroxyacyl-ACP dehydratase, UPF0336 protein Rv0637 | | Authors: | Singh, B.K, Biswas, R, Bhattacharyya, S, Basak, A, Das, A.K. | | Deposit date: | 2018-05-23 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | The C-terminal end of mycobacterial HadBC regulates AcpM interaction during the FAS-II pathway: a structural perspective.
Febs J., 2022
|
|
5DW8
 
 | |
6JQY
 
 | |
3V1T
 
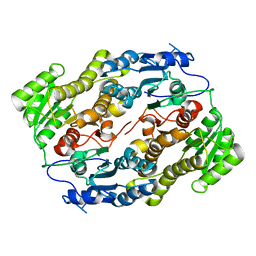 | |
4A04
 
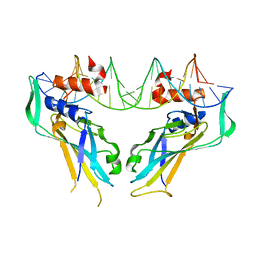 | | Structure of the DNA-bound T-box domain of human TBX1, a transcription factor associated with the DiGeorge syndrome | | Descriptor: | DNA, T-BOX TRANSCRIPTION FACTOR TBX1 | | Authors: | El Omari, K, De Mesmaeker, J, Karia, D, Ginn, H, Bhattacharya, S, Mancini, E.J. | | Deposit date: | 2011-09-07 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of the DNA-Bound T-Box Domain of Human Tbx1, a Transcription Factor Associated with the Digeorge Syndrome
Proteins, 80, 2012
|
|
3V1U
 
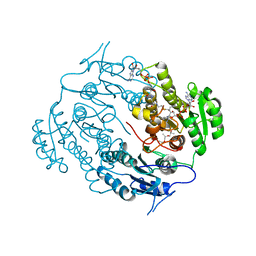 | | Crystal structure of a beta-ketoacyl reductase FabG4 from Mycobacterium tuberculosis H37Rv complexed with NAD+ and Hexanoyl-CoA at 2.5 Angstrom resolution | | Descriptor: | (2S,5R,8R,11S,14S,17S,21R)-5,8,11,14,17-PENTAMETHYL-4,7,10,13,16,19-HEXAOXADOCOSANE-2,21-DIOL, 3-oxoacyl-(Acyl-carrier-protein) reductase, HEXANOYL-COENZYME A, ... | | Authors: | Dutta, D, Bhattacharyya, S, Das, A.K. | | Deposit date: | 2011-12-10 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of hexanoyl-CoA bound to beta-ketoacyl reductase FabG4 of Mycobacterium tuberculosis
Biochem.J., 450, 2013
|
|
4PTK
 
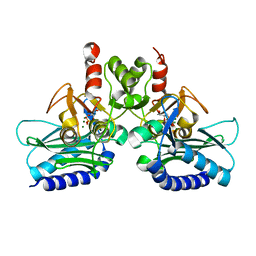 | | Crystal structure of Staphylococcal IMPase-I complex with 3Mg2+ and Phosphate | | Descriptor: | GLYCEROL, Inositol monophosphatase family protein, MAGNESIUM ION, ... | | Authors: | Dutta, A, Bhattacharyya, S, Dutta, D, Das, A.K. | | Deposit date: | 2014-03-11 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural elucidation of the binding site and mode of inhibition of Li(+) and Mg(2+) in inositol monophosphatase.
Febs J., 281, 2014
|
|
7XLY
 
 | | Crystal structure of FadA2 (Rv0243) from the fatty acid metabolic pathway of Mycobacterium tuberculosis | | Descriptor: | Probable acetyl-CoA acyltransferase FadA2 (3-ketoacyl-CoA thiolase) (Beta-ketothiolase), SULFATE ION | | Authors: | Singh, R, Kundu, P, Singh, B.K, Bhattacharyya, S, Das, A.K. | | Deposit date: | 2022-04-23 | | Release date: | 2023-04-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of FadA2 thiolase from Mycobacterium tuberculosis and prediction of its substrate specificity and membrane-anchoring properties.
Febs J., 290, 2023
|
|
4FW8
 
 | | Crystal structure of FABG4 complexed with Coenzyme NADH | | Descriptor: | (2S,5R,8R,11S,14S,17S,21R)-5,8,11,14,17-PENTAMETHYL-4,7,10,13,16,19-HEXAOXADOCOSANE-2,21-DIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-(Acyl-carrier-protein) reductase | | Authors: | Dutta, D, Bhattacharyya, S, Das, A.K. | | Deposit date: | 2012-06-30 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of hexanoyl-CoA bound to beta-ketoacyl reductase FabG4 of Mycobacterium tuberculosis
Biochem.J., 450, 2013
|
|
2RUO
 
 | | Solution Structure of Internal Fusion Peptide | | Descriptor: | UNP residues 873-888 of Spike glycoprotein | | Authors: | Mahajan, M, Bhattacharjya, S. | | Deposit date: | 2014-11-06 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structures and Localization of Potential Fusion Peptides and Pre-transmembrane Region of SARS-CoV: Implications in Membrane Fusion
To be Published
|
|
2RUM
 
 | | Solution structure of Fusion peptide | | Descriptor: | Fusion peptide of Spike glycoprotein | | Authors: | Mahajan, M, Bhattacharjya, S. | | Deposit date: | 2014-11-05 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structures and Localization of Potential Fusion Peptides and Pre-transmembrane Region of SARS-CoV: Implications in Membrane Fusion
To be Published
|
|
2RSW
 
 | |
2RUN
 
 | | Solution Structure of Pre Transmembrane domain | | Descriptor: | Pre-transmembrane domain of Spike glycoprotein | | Authors: | Mahajan, M, Bhattacharjya, S. | | Deposit date: | 2014-11-06 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structures and Localization of Potential Fusion Peptides and Pre-transmembrane Region of SARS-CoV: Implications in Membrane Fusion
To be Published
|
|
4I40
 
 | | crystal structure of Staphylococcal inositol monophosphatase-1: 50mM LiCl inhibited complex | | Descriptor: | GLYCEROL, Inositol monophosphatase family protein, MAGNESIUM ION, ... | | Authors: | Dutta, A, Bhattacharyya, S, Dutta, D, Das, A.K. | | Deposit date: | 2012-11-27 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural elucidation of the binding site and mode of inhibition of Li(+) and Mg(2+) in inositol monophosphatase.
Febs J., 281, 2014
|
|
4I3Y
 
 | | Crystal structure of Staphylococcal inositol monophosphatase-1: 100 mM LiCl soaked inhibitory complex | | Descriptor: | GLYCEROL, Inositol monophosphatase family protein, MAGNESIUM ION, ... | | Authors: | Dutta, A, Bhattacharyya, S, Dutta, D, Das, A.K. | | Deposit date: | 2012-11-26 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural elucidation of the binding site and mode of inhibition of Li(+) and Mg(2+) in inositol monophosphatase.
Febs J., 281, 2014
|
|
1XXG
 
 | |
1I8C
 
 | | SOLUTION STRUCTURE OF THE WATER-SOLUBLE FRAGMENT OF RAT HEPATIC APOCYTOCHROME B5 | | Descriptor: | CYTOCHROME B5 | | Authors: | Falzone, C.J, Wang, Y, Vu, B.C, Scott, N.L, Bhattacharya, S, Lecomte, J.T. | | Deposit date: | 2001-03-13 | | Release date: | 2001-05-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic perturbations induced by heme binding in cytochrome b5.
Biochemistry, 40, 2001
|
|
1I87
 
 | | SOLUTION STRUCTURE OF THE WATER-SOLUBLE FRAGMENT OF RAT HEPATIC APOCYTOCHROME B5 | | Descriptor: | CYTOCHROME B5 | | Authors: | Falzone, C.J, Wang, Y, Vu, B.C, Scott, N.L, Bhattacharya, S, Lecomte, J.T. | | Deposit date: | 2001-03-12 | | Release date: | 2001-05-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic perturbations induced by heme binding in cytochrome b5.
Biochemistry, 40, 2001
|
|
1LP3
 
 | | The Atomic Structure of Adeno-Associated Virus (AAV-2), a Vector for Human Gene Therapy | | Descriptor: | AAV-2 capsid protein | | Authors: | Xie, Q, Bu, W, Bhatia, S, Hare, J, Somasundaram, T, Azzi, A, Chapman, M.S. | | Deposit date: | 2002-05-07 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3Q6I
 
 | | Crystal structure of FabG4 and coenzyme binary complex | | Descriptor: | (2S,5R,8R,11S,14S,17S,21R)-5,8,11,14,17-PENTAMETHYL-4,7,10,13,16,19-HEXAOXADOCOSANE-2,21-DIOL, 3-oxoacyl-(Acyl-carrier-protein) reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dutta, D, Bhattacharyya, S, Das, A.K. | | Deposit date: | 2011-01-01 | | Release date: | 2012-01-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of holoFabG4
To be Published
|
|
3SJ7
 
 | |
2JOZ
 
 | | Solution NMR structure of protein yxeF, Northeast Structural Genomics Consortium target Sr500a | | Descriptor: | Hypothetical protein yxeF | | Authors: | Wu, Y, Liu, G, Zhang, Q, Bhatnagar, S, Chen, C, Nwosu, C, Xiao, R, Cunningham, K, Locke, J, Ma, L, Swapna, G, Baran, M, Acton, T, Montelione, G, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-04-10 | | Release date: | 2007-05-15 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein yxeF , Northeast Structural Genomics Consortium target Sr500a
To be Published
|
|
2K98
 
 | |
