4M6H
 
 | | Structure of the reduced, metal-free form of Mycobacterium tuberculosis peptidoglycan amidase Rv3717 | | Descriptor: | Peptidoglycan Amidase Rv3717 | | Authors: | Prigozhin, D.M, Mavrici, D, Huizar, J.P, Vansell, H.J, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural and Biochemical Analyses of Mycobacterium tuberculosis N-Acetylmuramyl-L-alanine Amidase Rv3717 Point to a Role in Peptidoglycan Fragment Recycling.
J.Biol.Chem., 288, 2013
|
|
3TGP
 
 | | Room temperature H-ras | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Fraser, J.S, Alber, T. | | Deposit date: | 2011-08-17 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3075 Å) | | Cite: | Accessing protein conformational ensembles using room-temperature X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4NX4
 
 | | Re-refinement of CAP-1 HIV-CA complex | | Descriptor: | 1-(3-chloro-4-methylphenyl)-3-{2-[({5-[(dimethylamino)methyl]-2-furyl}methyl)thio]ethyl}urea, CHLORIDE ION, Gag-Pol polyprotein, ... | | Authors: | Lang, P.T, Holton, J.M, Fraser, J.S, Alber, T. | | Deposit date: | 2013-12-08 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protein structural ensembles are revealed by redefining X-ray electron density noise.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4OGR
 
 | |
2O7A
 
 | | T4 lysozyme C-terminal fragment | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme | | Authors: | Echols, N, Kwon, E, Marqusee, S.M, Alber, T. | | Deposit date: | 2006-12-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Exploring subdomain cooperativity in T4 lysozyme I: Structural and energetic studies of a circular permutant and protein fragment.
Protein Sci., 16, 2007
|
|
1YWF
 
 | |
3OUN
 
 | |
3ORM
 
 | | Mycobacterium tuberculosis PknB kinase domain D76A mutant | | Descriptor: | MANGANESE (II) ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine protein kinase | | Authors: | Echols, N, Lombana, T.N, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-09-07 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric activation mechanism of the Mycobacterium tuberculosis receptor Ser/Thr protein kinase, PknB.
Structure, 18, 2010
|
|
3ORT
 
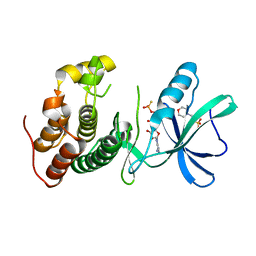 | | Mycobacterium tuberculosis PknB kinase domain L33D mutant (crystal form 6) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine protein kinase | | Authors: | Good, M.C, Echols, N, Lombana, T.N, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-09-07 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric activation mechanism of the Mycobacterium tuberculosis receptor Ser/Thr kinase, PknB
Structure, 18, 2010
|
|
3IP2
 
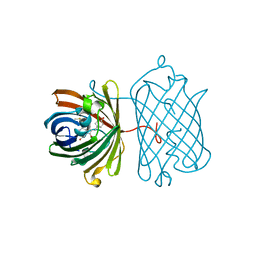 | | Crystal structure of red fluorescent protein Neptune at pH 7.0 | | Descriptor: | Neptune red fluorescent protein | | Authors: | Lin, M.Z, McKeown, M.R, Ng, H.L, Aguilera, T.A, Shaner, N.C, Ma, W, Adams, S.R, Campbell, R.E, Alber, T, Tsien, R.Y. | | Deposit date: | 2009-08-15 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Autofluorescent proteins with excitation in the optical window for intravital imaging in mammals.
Chem.Biol., 16, 2009
|
|
3ORI
 
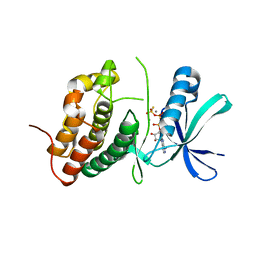 | | Mycobacterium tuberculosis PknB kinase domain L33D mutant (crystal form 1) | | Descriptor: | MANGANESE (II) ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine protein kinase | | Authors: | Lombana, T.N, Echols, N, Good, M.C, Thomsen, N.D, Ng, H.-L, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-09-07 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric activation mechanism of the Mycobacterium tuberculosis receptor Ser/Thr protein kinase, PknB.
Structure, 18, 2010
|
|
3OUK
 
 | |
3GOF
 
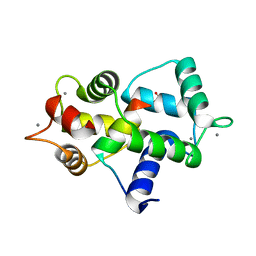 | | Calmodulin bound to peptide from macrophage nitric oxide synthase | | Descriptor: | CALCIUM ION, Calmodulin, Nitric oxide synthase, ... | | Authors: | Ng, H.L, Greenstein, A, Marletta, M, Wand, A.J, Alber, T. | | Deposit date: | 2009-03-19 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural diversity in calmodulin recognition of nitric oxide synthases
To be Published
|
|
3K0P
 
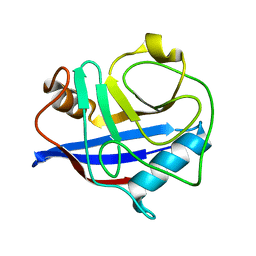 | |
3RMR
 
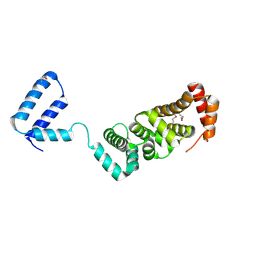 | |
3OUV
 
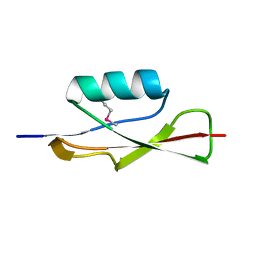 | |
3ORK
 
 | | Mycobacterium tuberculosis PknB kinase domain L33D mutant (crystal form 2) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Echols, N, Lombana, T.N, Thomsen, N.D, Ng, H.-L, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-09-07 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Allosteric activation mechanism of the Mycobacterium tuberculosis receptor Ser/Thr kinase, PknB
Structure, 18, 2010
|
|
3ORO
 
 | | Mycobacterium tuberculosis PknB kinase domain L33D mutant (crystal form 4) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine protein kinase | | Authors: | Good, M.C, Echols, N, Lombana, T.N, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-09-07 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric activation mechanism of the Mycobacterium tuberculosis receptor Ser/Thr protein kinase, PknB.
Structure, 18, 2010
|
|
3ORL
 
 | | Mycobacterium tuberculosis PknB kinase domain L33D mutant (crystal form 3) | | Descriptor: | MANGANESE (II) ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine protein kinase | | Authors: | Echols, N, Lombana, T.N, Thomsen, N.D, Ng, H.-L, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-09-07 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric activation mechanism of the Mycobacterium tuberculosis receptor Ser/Thr kinase, PknB
Structure, 18, 2010
|
|
3ORP
 
 | | Mycobacterium tuberculosis PknB kinase domain L33D mutant (crystal form 5) | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine protein kinase | | Authors: | Good, M.C, Echols, N, Lombana, T.N, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-09-07 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Allosteric activation mechanism of the Mycobacterium tuberculosis receptor Ser/Thr kinase, PknB
Structure, 18, 2010
|
|
3OTV
 
 | |
3OXH
 
 | | Mycobacterium tuberculosis kinase inhibitor homolog RV0577 | | Descriptor: | CHLORIDE ION, PARA-MERCURY-BENZENESULFONIC ACID, RV0577 PROTEIN, ... | | Authors: | Echols, N, Flynn, E.M, Stephenson, S, Ng, H.-L, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-09-21 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Biophysical Characterization of the Mycobacterium tuberculosis Protein Rv0577, a Protein Associated with Neutral Red Staining of Virulent Tuberculosis Strains and Homologue of the Streptomyces coelicolor Protein KbpA.
Biochemistry, 2017
|
|
2O79
 
 | | T4 lysozyme with C-terminal extension | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme, ... | | Authors: | Llinas, M, Crowder, S.M, Echols, N, Alber, T, Marqusee, S. | | Deposit date: | 2006-12-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring subdomain cooperativity in T4 lysozyme I: Structural and energetic studies of a circular permutant and protein fragment.
Protein Sci., 16, 2007
|
|
2OZ5
 
 | | Crystal structure of Mycobacterium tuberculosis protein tyrosine phosphatase PtpB in complex with the specific inhibitor OMTS | | Descriptor: | Phosphotyrosine protein phosphatase ptpb, {(3-CHLOROBENZYL)[(5-{[(3,3-DIPHENYLPROPYL)AMINO]SULFONYL}-2-THIENYL)METHYL]AMINO}(OXO)ACETIC ACID | | Authors: | Grundner, C, Gee, C.L, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Selective Inhibition of Mycobacterium tuberculosis Protein Tyrosine Phosphatase PtpB.
Structure, 15, 2007
|
|
2O5G
 
 | | Calmodulin-smooth muscle light chain kinase peptide complex | | Descriptor: | CALCIUM ION, Calmodulin, SULFATE ION, ... | | Authors: | Valentine, K.G, Ng, H.L, Schneeweis, J.K, Kranz, J.K, Frederick, K.K, Alber, T, Wand, A.J. | | Deposit date: | 2006-12-05 | | Release date: | 2007-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Ultrahigh resolution crystal structure of calmodulin-smooth muscle light kinase peptide complex
To be Published
|
|
