6OSY
 
 | |
6L5M
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5N
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-unwound state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5L
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at apo state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5O
 
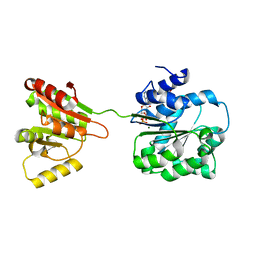 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-hydrolysis state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
8T5C
 
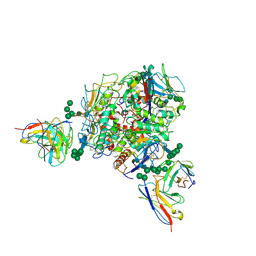 | | Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8.11G Heavy Chain, 8.11G Light Chain, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cleavage-intermediate Lassa virus trimer elicits neutralizing responses, identifies neutralizing nanobodies, and reveals an apex-situated site-of-vulnerability.
Nat Commun, 15, 2024
|
|
6OT1
 
 | |
8AHH
 
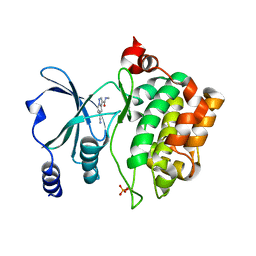 | |
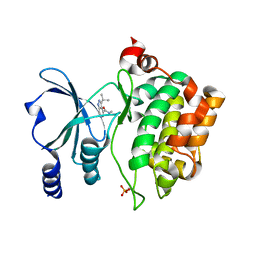
8AHG
 
 | |
8AHE
 
 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | SULFATE ION, UDP-N-acetylglucosamine 2-epimerase, ~{N},5-dimethyl-1-phenyl-pyrazole-4-sulfonamide | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
8AHF
 
 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | (2~{R},4~{S})-4-[bis(fluoranyl)methoxy]-~{N}-methyl-1-(2~{H}-pyrazolo[4,3-b]pyridin-6-ylcarbonyl)pyrrolidine-2-carboxamide, SULFATE ION, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.271 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
8AHI
 
 | |
5TKJ
 
 | | Structure of vaccine-elicited diverse HIV-1 neutralizing antibody vFP1.01 in complex with HIV-1 fusion peptide residue 512-519 | | Descriptor: | HIV-1 fusion peptide residue 512-519, SULFATE ION, vFP1.01 chimeric mouse antibody heavy chain, ... | | Authors: | Xu, K, Liu, K, Kwong, P.D. | | Deposit date: | 2016-10-06 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.118 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
5TKK
 
 | |
9B97
 
 | | Crystal structure of the human PAD2 protein bound to small molecule | | Descriptor: | (1P)-N~3'~-[(2S)-3-cyclohexyl-1-(methylamino)-1-oxopropan-2-yl]-N~3~,N~3~-diethyl-4-fluoro-5'-{[4-(4-phenylbutyl)piperazin-1-yl]methyl}[1,1'-biphenyl]-3,3'-dicarboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Byrnes, L.J, Vajdos, F. | | Deposit date: | 2024-04-01 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery, Characterization, and Structure of a Cell Active PAD2 Inhibitor Acting through a Novel Allosteric Mechanism.
Acs Chem.Biol., 19, 2024
|
|
9B96
 
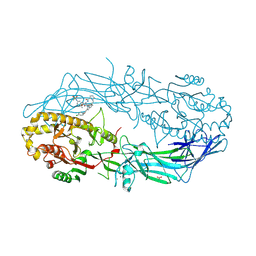 | | Crystal structure of the human PAD2 protein bound to inhibitor | | Descriptor: | 1-({(2P)-1-{(1R)-1-(2-bromophenyl)-3-[5-(methanesulfonamido)-2-methylanilino]-3-oxopropyl}-2-[3-(4-chlorophenoxy)phenyl]-1H-1,3-benzimidazol-6-yl}methyl)-N-methyl-D-prolinamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Byrnes, L.J, Vajdos, F. | | Deposit date: | 2024-04-01 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery, Characterization, and Structure of a Cell Active PAD2 Inhibitor Acting through a Novel Allosteric Mechanism.
Acs Chem.Biol., 19, 2024
|
|
9B98
 
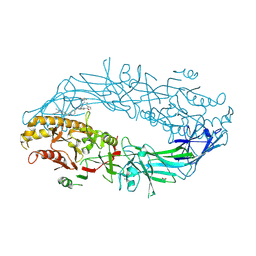 | | Crystal structure of the human PAD2 protein bound to small molecule | | Descriptor: | (5P)-N,N-diethyl-2-fluoro-5-(2-[({1-[2-(methylamino)-2-oxoethyl]cyclohexyl}methyl)amino]-6-{methyl[1-(2-methyl-1-phenyl-1H-1,3-benzimidazole-5-carbonyl)piperidin-4-yl]amino}pyrimidin-4-yl)benzamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Byrnes, L.J, Vajdos, F. | | Deposit date: | 2024-04-01 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Discovery, Characterization, and Structure of a Cell Active PAD2 Inhibitor Acting through a Novel Allosteric Mechanism.
Acs Chem.Biol., 19, 2024
|
|
2P6B
 
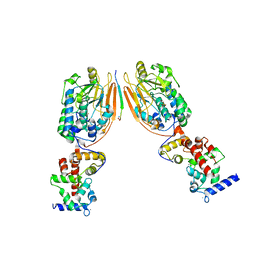 | | Crystal Structure of Human Calcineurin in Complex with PVIVIT Peptide | | Descriptor: | CALCIUM ION, Calcineurin subunit B isoform 1, Calmodulin-dependent calcineurin A subunit alpha isoform, ... | | Authors: | Li, H, Zhang, L, Rao, A, Harrison, S.C, Hogan, P.G. | | Deposit date: | 2007-03-16 | | Release date: | 2007-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of calcineurin in complex with PVIVIT peptide: Portrait of a low-affinity signalling interaction
J.Mol.Biol., 369, 2007
|
|
6CDO
 
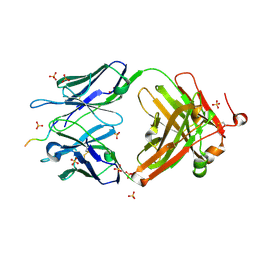 | | Structure of vaccine-elicited HIV-1 neutralizing antibody vFP16.02 in complex with HIV-1 fusion peptide residue 512-519 | | Descriptor: | HIV-1 fusion peptide 512-519, SULFATE ION, vFP16.02 Fab heavy chain, ... | | Authors: | Xu, K, Liu, K, Kwong, P.D. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-16 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
6CDP
 
 | | Vaccine-elicited HIV-1 neutralizing antibody vFP20.01 in complex with HIV-1 fusion peptide residue 512-519 | | Descriptor: | HIV-1 fusion peptide 512-519, SULFATE ION, vFP20.01 Fab heavy chain, ... | | Authors: | Xu, K, Liu, K, Kwong, P.D. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.456 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
6CDE
 
 | | Cryo-EM structure at 3.8 A resolution of vaccine-elicited antibody vFP20.01 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein 120, ... | | Authors: | Acharya, P, Xu, K, Liu, K, Carragher, B, Potter, C.S, Kwong, P.D. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-16 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
6CDI
 
 | | Cryo-EM structure at 3.6 A resolution of vaccine-elicited antibody vFP16.02 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein 120, ... | | Authors: | Acharya, P, Xu, K, Liu, K, Carragher, B, Potter, C.S, Kwong, P.D. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-16 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
6CDM
 
 | |
5X4G
 
 | | Crystal structure of Fab fragment of anti-CD147 monoclonal antibody 6H8 | | Descriptor: | 6H8 Fab heavy chain, 6H8 Fab light chain | | Authors: | Lin, P, Zhang, M.-Y, Chen, X, Ye, S, Yu, X.-L, Zhang, R.-G, Zhu, P, Chen, Z.-N. | | Deposit date: | 2017-02-13 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of CD147 C2 domain in complex with Fab of its monoclonal antibody
To Be Published
|
|
5WYL
 
 | |
