4HK5
 
 | | Crystal structure of Cordyceps militaris IDCase in apo form | | Descriptor: | Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Zhu, J, Ding, J. | | Deposit date: | 2012-10-15 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
5YRN
 
 | | Structure of RIP2 CARD domain | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Wu, B, Gong, Q. | | Deposit date: | 2017-11-09 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of RIP2 activation and signaling.
Nat Commun, 9, 2018
|
|
2QE3
 
 | | Crystal structure of human tl1a extracellular domain | | Descriptor: | CHLORIDE ION, TNF superfamily ligand TL1A | | Authors: | Zhan, C, Yan, Q, Patskovsky, Y, Shi, W, Toro, R, Bonanno, J, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-06-22 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical and structural characterization of the human TL1A ectodomain.
Biochemistry, 48, 2009
|
|
2P0H
 
 | | ArhGAP9 PH domain in complex with Ins(1,3,4)P3 | | Descriptor: | (1S,3S,4S)-1,3,4-TRIPHOSPHO-MYO-INOSITOL, Rho GTPase-activating protein 9 | | Authors: | Ceccarelli, D.F.J, Blasutig, I, Goudreault, M, Ruston, J, Pawson, T, Sicheri, F. | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Non-canonical Interaction of Phosphoinositides with Pleckstrin Homology Domains of Tiam1 and ArhGAP9.
J.Biol.Chem., 282, 2007
|
|
2OCA
 
 | | The crystal structure of T4 UvsW | | Descriptor: | ATP-dependent DNA helicase uvsW | | Authors: | Kerr, I.D, White, S.W. | | Deposit date: | 2006-12-20 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic and NMR Analyses of UvsW and UvsW.1 from Bacteriophage T4.
J.Biol.Chem., 282, 2007
|
|
4HK6
 
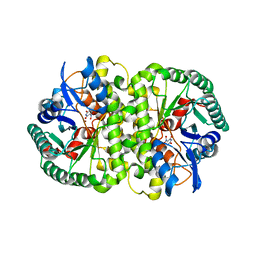 | |
6Y09
 
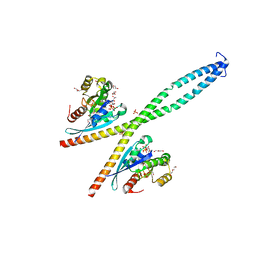 | |
2P0F
 
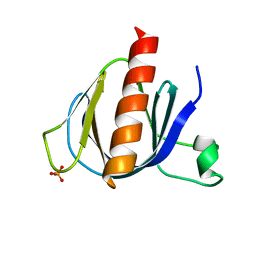 | | ArhGAP9 PH domain in complex with Ins(1,3,5)P3 | | Descriptor: | PHOSPHATE ION, Rho GTPase-activating protein 9 | | Authors: | Ceccarelli, D.F.J, Blasutig, I, Goudreault, M, Ruston, J, Pawson, T, Sicheri, F. | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Non-canonical Interaction of Phosphoinositides with Pleckstrin Homology Domains of Tiam1 and ArhGAP9.
J.Biol.Chem., 282, 2007
|
|
2P0D
 
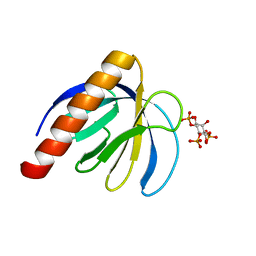 | | ArhGAP9 PH domain in complex with Ins(1,4,5)P3 | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Rho GTPase-activating protein 9 | | Authors: | Ceccarelli, D.F.J, Blasutig, I, Goudreault, M, Ruston, J, Pawson, T, Sicheri, F. | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Non-canonical Interaction of Phosphoinositides with Pleckstrin Homology Domains of Tiam1 and ArhGAP9.
J.Biol.Chem., 282, 2007
|
|
7WK2
 
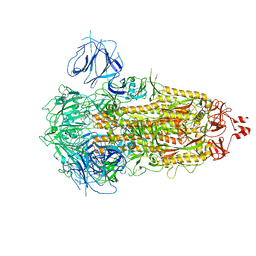 | | SARS-CoV-2 Omicron S-close | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-01-08 | | Release date: | 2022-01-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
7WVO
 
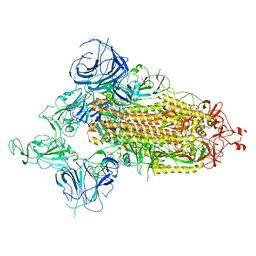 | | SARS-CoV-2 Omicron S-open-2 | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2022-03-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
7WVN
 
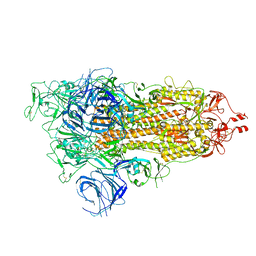 | | SARS-CoV-2 Omicron S-open | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2022-03-02 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
2R9C
 
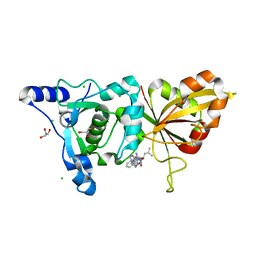 | | Calpain 1 proteolytic core inactivated by ZLAK-3001, an alpha-ketoamide | | Descriptor: | CALCIUM ION, CHLORIDE ION, Calpain-1 catalytic subunit, ... | | Authors: | Qian, J, Campbell, R.L, Davies, P.L. | | Deposit date: | 2007-09-12 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cocrystal structures of primed side-extending alpha-ketoamide inhibitors reveal novel calpain-inhibitor aromatic interactions.
J.Med.Chem., 51, 2008
|
|
2R9F
 
 | | Calpain 1 proteolytic core inactivated by ZLAK-3002, an alpha-ketoamide | | Descriptor: | CALCIUM ION, CHLORIDE ION, Calpain-1 catalytic subunit, ... | | Authors: | Qian, J, Campbell, R.L, Davies, P.L. | | Deposit date: | 2007-09-12 | | Release date: | 2008-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cocrystal structures of primed side-extending alpha-ketoamide inhibitors reveal novel calpain-inhibitor aromatic interactions.
J.Med.Chem., 51, 2008
|
|
2RJK
 
 | | Crystal Structure of Human TL1A Extracellular Domain C95S Mutant | | Descriptor: | TNF superfamily ligand TL1A | | Authors: | Zhan, C, Yan, Q, Patskovsky, Y, Shi, W, Ramagopal, U.A, Toro, R, Bonanno, J, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-10-15 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and structural characterization of the human TL1A ectodomain.
Biochemistry, 48, 2009
|
|
4IT7
 
 | | Crystal structure of Al-CPI | | Descriptor: | CPI | | Authors: | Mei, G.Q, Liu, S.L, Sun, M.Z, Liu, J. | | Deposit date: | 2013-01-17 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Immunomodulatory Function of Cysteine Protease Inhibitor from Human Roundworm Ascaris lumbricoides.
Plos One, 9, 2014
|
|
2RJL
 
 | | Crystal structure of human TL1A extracellular domain C95S/C135S mutant | | Descriptor: | TNF superfamily ligand TL1A | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Shi, W, Toro, R, Bonanno, J, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-10-15 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biochemical and structural characterization of the human TL1A ectodomain.
Biochemistry, 48, 2009
|
|
4HJW
 
 | | Crystal structure of Metarhizium anisopliae IDCase in apo form | | Descriptor: | Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Zhu, J, Ding, J. | | Deposit date: | 2012-10-14 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
7VDL
 
 | |
7VDH
 
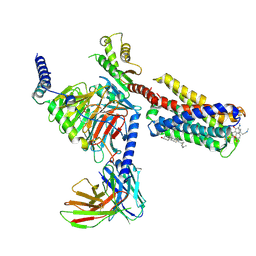 | | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with C48/80, state2 | | Descriptor: | 2-[4-methoxy-3-[[2-methoxy-3-[[2-methoxy-5-[2-(methylamino)ethyl]phenyl]methyl]-5-[2-(methylamino)ethyl]phenyl]methyl]phenyl]-~{N}-methyl-ethanamine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, Y, Yang, F. | | Deposit date: | 2021-09-07 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure, function and pharmacology of human itch receptor complexes.
Nature, 600, 2021
|
|
7VDM
 
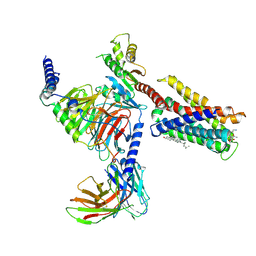 | | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with substance P | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, Y, Yang, F. | | Deposit date: | 2021-09-07 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structure, function and pharmacology of human itch receptor complexes.
Nature, 600, 2021
|
|
7XVQ
 
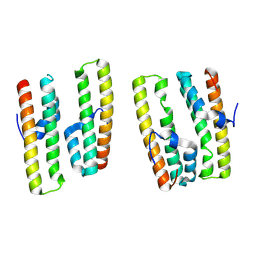 | | Crystal structure of AcrIIC4 | | Descriptor: | Uncharacterized protein | | Authors: | Zhang, H, Li, X.Z, Song, G.Y. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-09 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibitory mechanism of CRISPR-Cas9 by AcrIIC4.
Nucleic Acids Res., 51, 2023
|
|
5OOG
 
 | | Human biliverdin IX beta reductase: NADP/Phloxine B ternary complex | | Descriptor: | Flavin reductase (NADPH), GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Manso, J.A, Pereira, P.J.B. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | In silicoand crystallographic studies identify key structural features of biliverdin IX beta reductase inhibitors having nanomolar potency.
J. Biol. Chem., 293, 2018
|
|
7VUZ
 
 | | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with PAMP-12, state2 | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, Y, Yang, F. | | Deposit date: | 2021-11-04 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure, function and pharmacology of human itch receptor complexes.
Nature, 600, 2021
|
|
7VV5
 
 | | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with C48/80, state1 | | Descriptor: | 2-[4-methoxy-3-[[2-methoxy-3-[[2-methoxy-5-[2-(methylamino)ethyl]phenyl]methyl]-5-[2-(methylamino)ethyl]phenyl]methyl]phenyl]-~{N}-methyl-ethanamine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, Y, Yang, F. | | Deposit date: | 2021-11-04 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structure, function and pharmacology of human itch receptor complexes.
Nature, 600, 2021
|
|
