8V8K
 
 | | Crystal Structure of Nanobody NbE | | Descriptor: | Nanobody NbE | | Authors: | Koehl, A, Manglik, A, Yu, J, Kumar, A, Zhang, X, Martin, C, Raia, P, Steyaert, J, Ballet, S, Boland, A, Stoeber, M. | | Deposit date: | 2023-12-05 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis of mu-Opioid Receptor-Targeting by a Nanobody Antagonist
To Be Published
|
|
8W9E
 
 | |
8W9F
 
 | |
8WOQ
 
 | | Cryo-EM structure of human SIDT1 protein with C1 symmetry at neutral pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Liu, W, Tang, M, Wang, J, Zhang, X, Wu, S, Ru, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1.
Cell Discov, 10, 2024
|
|
8WOT
 
 | | Cryo-EM structure of human SIDT1 protein with C2 symmetry at low pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Liu, W, Tang, M, Wang, J, Zhang, X, Wu, S, Ru, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1.
Cell Discov, 10, 2024
|
|
8WOS
 
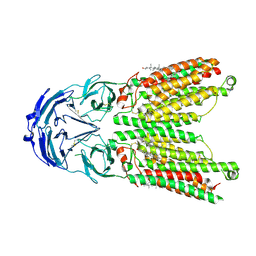 | | Cryo-EM structure of human SIDT1 protein with C1 symmetry at low pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Liu, W, Tang, M, Wang, J, Zhang, X, Wu, S, Ru, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1.
Cell Discov, 10, 2024
|
|
8WOR
 
 | | Cryo-EM structure of human SIDT1 protein with C2 symmetry at neutral pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Liu, W, Tang, M, Wang, J, Zhang, X, Wu, S, Ru, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1.
Cell Discov, 10, 2024
|
|
8W9D
 
 | |
8W9C
 
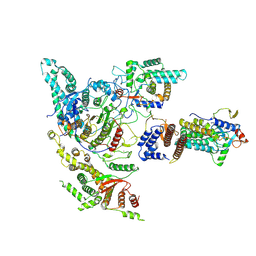 | |
5YC6
 
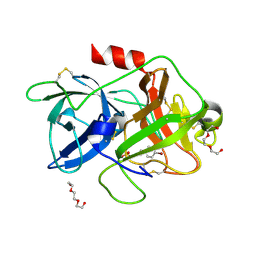 | | The crystal structure of uPA in complex with 4-Bromobenzylamirne at pH4.6 | | Descriptor: | 1-(4-BROMOPHENYL)METHANAMINE, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Jiang, L.G, Zhang, X, Huang, M.D. | | Deposit date: | 2017-09-06 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Halogen bonding for the design of inhibitors by targeting the S1 pocket of serine proteases
Rsc Adv, 8, 2018
|
|
7EGK
 
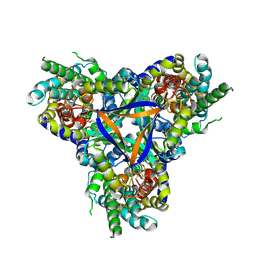 | | Bicarbonate transporter complex SbtA-SbtB bound to AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Membrane-associated protein SbtB, SODIUM ION, ... | | Authors: | Fang, S, Huang, X, Zhang, X, Zhang, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular mechanism underlying transport and allosteric inhibition of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EGL
 
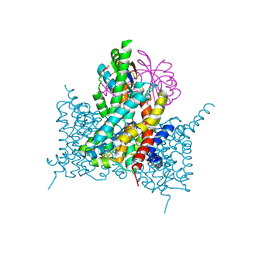 | | Bicarbonate transporter complex SbtA-SbtB bound to HCO3- | | Descriptor: | BICARBONATE ION, Membrane-associated protein SbtB, SODIUM ION, ... | | Authors: | Fang, S, Huang, X, Zhang, X, Zhang, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular mechanism underlying transport and allosteric inhibition of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5YC7
 
 | |
5ZGC
 
 | |
6GSC
 
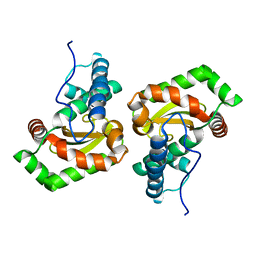 | | Sphingobacterium sp. T2 manganese superoxide dismutase catalyses the oxidative demethylation of polymeric lignin via generation of hydroxyl radical | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Rashid, G.M.M, Zhang, X, Wilkinson, R.C, Fulop, V, Cottyn, B, Baumberger, S, Bugg, T.D.H. | | Deposit date: | 2018-06-13 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Sphingobacterium sp. T2 Manganese Superoxide Dismutase Catalyzes the Oxidative Demethylation of Polymeric Lignin via Generation of Hydroxyl Radical.
ACS Chem. Biol., 13, 2018
|
|
7FAU
 
 | | Structure Determination of the NB1B11-RBD Complex | | Descriptor: | NB_1B11, Spike protein S1, ZINC ION | | Authors: | Shi, Z.Z, Li, X.X, Wang, L, Sun, Z.C, Zhang, H.W, Chen, X.C, Cui, Q.Q, Qiao, H.R, Lan, Z.Y, Zhang, X. | | Deposit date: | 2021-07-07 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of nanobodies neutralizing SARS-CoV-2 variants.
Structure, 30, 2022
|
|
8XKV
 
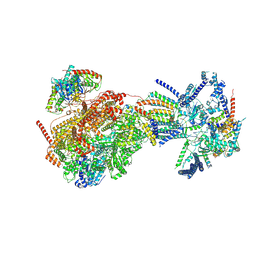 | | Cryo-EM structure of the Ycf2-FtsHi motor complex from Arabidopsis in Apo state | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, ATP-dependent zinc metalloprotease FTSH 12, chloroplastic, ... | | Authors: | Liang, K, Zhan, X, Xu, Q, Wu, J, Yan, Z. | | Deposit date: | 2023-12-25 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the chloroplast protein import in land plants.
Cell, 2024
|
|
8XKU
 
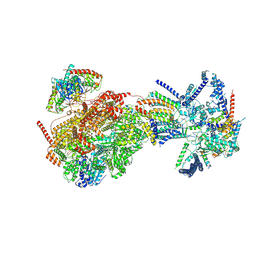 | | Cryo-EM structure of the Ycf2-FtsHi motor complex from Arabidopsis in ATP-bound state | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent zinc metalloprotease FTSH 12, ... | | Authors: | Liang, K, Zhan, X, Xu, Q, Wu, J, Yan, Z. | | Deposit date: | 2023-12-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the chloroplast protein import in land plants.
Cell, 2024
|
|
5Z1C
 
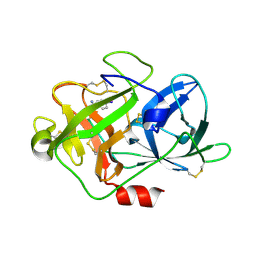 | | The crystal structure of uPA in complex with 4-Iodobenzylamine at pH7.4 | | Descriptor: | 1-(4-iodophenyl)methanamine, Urokinase-type plasminogen activator | | Authors: | Jiang, L.G, Zhang, X, Luo, Z.P, Huang, M.D. | | Deposit date: | 2017-12-25 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Halogen bonding for the design of inhibitors by targeting the S1 pocket of serine proteases
Rsc Adv, 49, 2018
|
|
7WQR
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound 28 | | Descriptor: | Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ~{N}-(3-chlorophenyl)-2-[(3,5-dimethyl-1,2-oxazol-4-yl)methoxy]benzamide | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Sun, H, Fang, P. | | Deposit date: | 2022-01-25 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.124 Å) | | Cite: | Development of highly potent and specific AKR1C3 inhibitors to restore the chemosensitivity of drug-resistant breast cancer.
Eur.J.Med.Chem., 247, 2022
|
|
7WQS
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound 25 | | Descriptor: | 2-[(3,5-dimethyl-1,2-oxazol-4-yl)methoxy]-~{N}-(3-methoxyphenyl)benzamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Sun, H, Fang, P. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Development of highly potent and specific AKR1C3 inhibitors to restore the chemosensitivity of drug-resistant breast cancer.
Eur.J.Med.Chem., 247, 2022
|
|
7WQM
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound 24 | | Descriptor: | 2-[(3,5-dimethyl-1,2-oxazol-4-yl)methoxy]-~{N}-(2-methoxyphenyl)benzamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Sun, H, Fang, P. | | Deposit date: | 2022-01-25 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Development of highly potent and specific AKR1C3 inhibitors to restore the chemosensitivity of drug-resistant breast cancer.
Eur.J.Med.Chem., 247, 2022
|
|
6IPO
 
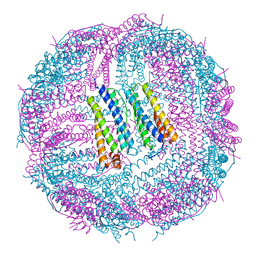 | | Ferritin mutant C90A/C102A/C130A/D144C | | Descriptor: | Ferritin heavy chain, MAGNESIUM ION | | Authors: | Zang, J, Chen, H, Zhang, X, Zhao, G. | | Deposit date: | 2018-11-03 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Disulfide-mediated conversion of 8-mer bowl-like protein architecture into three different nanocages.
Nat Commun, 10, 2019
|
|
2L37
 
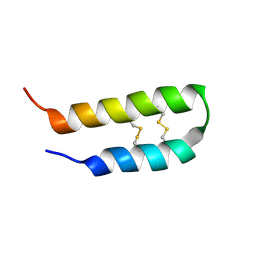 | | 3D solution structure of arginine/glutamate-rich polypeptide Luffin P1 from the seeds of sponge gourd (Luffa cylindrical) | | Descriptor: | Ribosome-inactivating protein luffin P1 | | Authors: | Ng, Y.M, Yang, Y, Sze, K.H, Zhang, X, Zheng, Y.T, Shaw, P.C. | | Deposit date: | 2010-09-08 | | Release date: | 2011-01-19 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structural characterization and anti-HIV-1 activities of arginine/glutamate-rich polypeptide Luffin P1 from the seeds of sponge gourd (Luffa cylindrical).
J.Struct.Biol., 2010
|
|
7VH3
 
 | |
