7R3L
 
 | | PARP14 catalytic domain in complex with OUL40 | | Descriptor: | 6-methyl-[1,2,4]triazolo[3,4-b][1,3]benzothiazole, CHLORIDE ION, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Maksimainen, M.M, Murthy, S, Lehtio, L. | | Deposit date: | 2022-02-07 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
7R3O
 
 | | PARP15 catalytic domain in complex with OUL40 | | Descriptor: | 6-methyl-[1,2,4]triazolo[3,4-b][1,3]benzothiazole, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Murthy, S, Lehtio, L. | | Deposit date: | 2022-02-07 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
7R4A
 
 | | PARP15 catalytic domain in complex with OUL188 | | Descriptor: | 6,8-dimethyl-[1,2,4]triazolo[3,4-b][1,3]benzothiazole, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Murthy, S, Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2022-02-08 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
1N65
 
 | |
7R5X
 
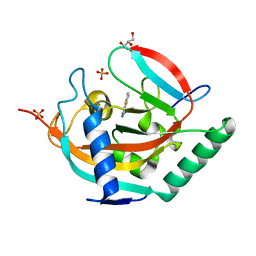 | | Tankyrase 2 in complex with an inhibitor (OUL211) | | Descriptor: | 7-methyl-[1,2,4]triazolo[3,4-b][1,3]benzothiazole, GLYCEROL, Poly [ADP-ribose] polymerase tankyrase-2, ... | | Authors: | Sowa, S.T, Lehtio, L. | | Deposit date: | 2022-02-11 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
1N6J
 
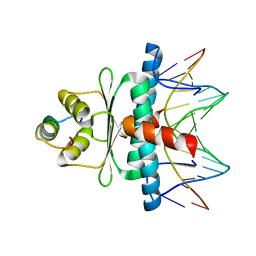 | | Structural basis of sequence-specific recruitment of histone deacetylases by Myocyte Enhancer Factor-2 | | Descriptor: | 5'-D(*AP*GP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*GP*C)-3', 5'-D(*GP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*CP*T)-3', Calcineurin-binding protein Cabin 1, ... | | Authors: | Han, A, Pan, F, Stroud, J.C, Youn, H.D, Liu, J.O, Chen, L. | | Deposit date: | 2002-11-11 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sequence-specific recruitment of transcriptional co-repressor Cabin1 by myocyte enhancer factor-2
Nature, 422, 2003
|
|
5T5H
 
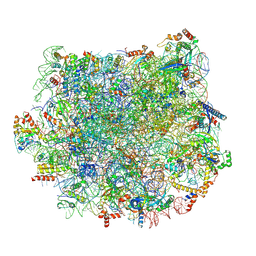 | | Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit | | Descriptor: | 40S ribosomal protein L14, 5.8S rRNA, 5S rRNA, ... | | Authors: | Liu, Z, Gutierrez-Vargas, C, Wei, J, Grassucci, R.A, Ramesh, M, Espina, N, Sun, M, Tutuncuoglu, B, Madison-Antenucci, S, Woolford Jr, J.L, Tong, L, Frank, J. | | Deposit date: | 2016-08-31 | | Release date: | 2016-10-12 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5SZO
 
 | | Protocadherin Gamma B7 extracellular cadherin domains 1-4 P41212 crystal form | | Descriptor: | CALCIUM ION, Protocadherin Gamma B7, alpha-D-mannopyranose, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2016-08-14 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.612 Å) | | Cite: | gamma-Protocadherin structural diversity and functional implications.
Elife, 5, 2016
|
|
5T48
 
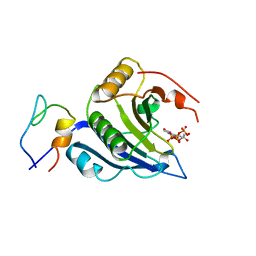 | | Crystal structure of the D. melanogaster eIF4E-eIF4G complex without lateral contact | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4G, ... | | Authors: | Gruener, S, Peter, D, Weber, R, Wohlbold, L, Chung, M.-Y, Weichenrieder, O, Valkov, E, Igreja, C, Izaurralde, E. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Structures of eIF4E-eIF4G Complexes Reveal an Extended Interface to Regulate Translation Initiation.
Mol.Cell, 64, 2016
|
|
1N72
 
 | | Structure and Ligand of a Histone Acetyltransferase Bromodomain | | Descriptor: | HISTONE ACETYLTRANSFERASE | | Authors: | Dhalluin, C, Carlson, J.E, Zeng, L, He, C, Aggarwal, A.K, Zhou, M.-M. | | Deposit date: | 2002-11-12 | | Release date: | 2002-12-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and Ligand of a Histone Acetyltransferase Bromodomain
Nature, 399, 1999
|
|
1N70
 
 | |
1LA1
 
 | | Gro-EL Fragment (Apical Domain) Comprising Residues 188-379 | | Descriptor: | GroEL | | Authors: | Ashcroft, A.E, Brinker, A, Coyle, J.E, Weber, F, Kaiser, M, Moroder, L, Parsons, M.R, Jager, J, Hartl, U.F, Hayer-Hartl, M, Radford, S.E. | | Deposit date: | 2002-03-27 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural plasticity and noncovalent substrate binding in the GroEL apical domain. A study using electrospay ionization mass spectrometry and fluorescence binding studies.
J.Biol.Chem., 277, 2002
|
|
4CDQ
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP2 | | Descriptor: | 4-((5-(2-oxo-3-(pyridin-4-yl)imidazolidin-1-yl)pentyl)oxy)benzaldehyde O-ethyl oxime, SODIUM ION, VP1, ... | | Authors: | DeColibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-05 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
1NC6
 
 | | Potent, small molecule inhibitors of human mast cell tryptase. Anti-asthmatic action of a dipeptide-based transition state analogue containing benzothiazole ketone | | Descriptor: | (2S,4R)-1-ACETYL-N-[(1S)-4-[(AMINOIMINOMETHYL)AMINO]-1-(2-BENZOTHIAZOLYLCARBONYL)BUTYL]-4-HYDROXY-2-PYRROLIDINECARBOXAMIDE, CALCIUM ION, SULFATE ION, ... | | Authors: | Costanzo, M.J, Yabut, S.C, Almond Jr, H.R, Andrade-Gordon, P, Corcoran, T.W, de Garavilla, L, Kauffman, J.A, Abraham, W.M, Recacha, R, Chattopadhyay, D, Maryanoff, B.E. | | Deposit date: | 2002-12-04 | | Release date: | 2003-09-23 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Potent, Small-Molecule Inhibitors of Human Mast Cell Tryptase. Antiasthmatic Action of a Dipeptide-Based Transition-State Analogue Containing a Benzothiazole Ketone.
J.Med.Chem., 46, 2003
|
|
1N9X
 
 | | structure of microgravity-grown oxidized myoglobin mutant YQR (ISS8A) | | Descriptor: | HYDROXIDE ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Miele, A.E, Sciara, G, Federici, L, Draghi, F, Brunori, M, Vallone, B. | | Deposit date: | 2002-11-26 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analysis of the effect of microgravity on protein crystal quality: the case of a myoglobin triple mutant.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1ND5
 
 | | Crystal Structures of Human Prostatic Acid Phosphatase in Complex with a Phosphate Ion and alpha-Benzylaminobenzylphosphonic Acid Update the Mechanistic Picture and Offer New Insights into Inhibitor Design | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ortlund, E, LaCount, M.W, Lebioda, L. | | Deposit date: | 2002-12-07 | | Release date: | 2002-12-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of human prostatic acid phosphatase in complex with a
phosphate ion and alpha-benzylaminobenzylphosphonic acid update the
mechanistic picture and offer new insights into inhibitor design
Biochemistry, 42, 2003
|
|
1LEK
 
 | | Crystal Structure of H-2Kbm3 bound to dEV8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Luz, J.G, Huang, M, Garcia, K.C, Rudolph, M.G, Apostolopoulos, V, Teyton, L, Wilson, I.A. | | Deposit date: | 2002-04-09 | | Release date: | 2002-06-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural comparison of allogeneic and syngeneic T cell receptor-peptide-major histocompatibility complex complexes: a buried alloreactive mutation subtly alters peptide presentation substantially increasing V(beta) Interactions.
J.Exp.Med., 195, 2002
|
|
1KGM
 
 | | SOLUTION STRUCTURE OF THE SMALL SERINE PROTEASE INHIBITOR SGCI | | Descriptor: | SERINE PROTEASE INHIBITOR I | | Authors: | Gaspari, Z, Patthy, A, Graf, L, Perczel, A. | | Deposit date: | 2001-11-28 | | Release date: | 2001-12-12 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Comparative structure analysis of proteinase inhibitors from the desert locust, Schistocerca gregaria.
Eur.J.Biochem., 269, 2002
|
|
1NDF
 
 | | Carnitine Acetyltransferase in Complex with Carnitine | | Descriptor: | CARNITINE, Carnitine Acetyltransferase | | Authors: | Jogl, G, Tong, L. | | Deposit date: | 2002-12-09 | | Release date: | 2003-01-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Carnitine Acetyltransferase and Implications for the Catalytic Mechanism and Fatty Acid Transport
Cell(Cambridge,Mass.), 112, 2003
|
|
1KFR
 
 | | Structural plasticity in the eight-helix fold of a trematode hemoglobin | | Descriptor: | Hemoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Milani, M, Pesce, A, Dewilde, S, Ascenzi, P, Moens, L, Bolognesi, M. | | Deposit date: | 2001-11-22 | | Release date: | 2002-04-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural plasticity in the eight-helix fold of a trematode haemoglobin.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5T1A
 
 | | Structure of CC Chemokine Receptor 2 with Orthosteric and Allosteric Antagonists | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{R})-1-(4-chloranyl-2-fluoranyl-phenyl)-2-cyclohexyl-3-ethanoyl-4-oxidanyl-2~{H}-pyrrol-5-one, (3S)-1-{(1S,2R,4R)-4-[methyl(propan-2-yl)amino]-2-propylcyclohexyl}-3-{[6-(trifluoromethyl)quinazolin-4-yl]amino}pyrrolidin-2-one, ... | | Authors: | Zheng, Y, Qin, L, Ortiz Zacarias, N.V, de Vries, H, Han, G.W, Gustavsson, M, Dabros, M, Zhao, C, Cherney, R.J, Carter, P, Stamos, D, Abagyan, R, Cherezov, V, Stevens, R.C, IJzerman, A.P, Heitman, L.H, Tebben, A, Kufareva, I, Handel, T.M. | | Deposit date: | 2016-08-18 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists.
Nature, 540, 2016
|
|
5T5S
 
 | | A fragment of a human tRNA synthetase | | Descriptor: | Alanine--tRNA ligase, cytoplasmic | | Authors: | Sun, L, Schimmel, P. | | Deposit date: | 2016-08-31 | | Release date: | 2016-11-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Two crystal structures reveal design for repurposing the C-Ala domain of human AlaRS.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
1KGL
 
 | | Solution structure of cellular retinol binding protein type-I in complex with all-trans-retinol | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN TYPE I, RETINOL | | Authors: | Franzoni, L, Luecke, C, Perez, C, Cavazzini, D, Rademacher, M, Ludwig, C, Spisni, A, Rossi, G.L, Rueterjans, H. | | Deposit date: | 2001-11-27 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and Backbone Dynamics of Apo- and Holo-cellular Retinol-binding
Protein in Solution.
J.Biol.Chem., 277, 2002
|
|
1NXU
 
 | | CRYSTAL STRUCTURE OF E. COLI HYPOTHETICAL OXIDOREDUCTASE YIAK NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ER82. | | Descriptor: | Hypothetical oxidoreductase yiaK, SULFATE ION | | Authors: | Forouhar, F, Lee, I, Benach, J, Kulkarni, K, Xiao, R, Acton, T.B, Shastry, R, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel NAD-binding Protein Revealed by the Crystal Structure of 2,3-Diketo-L-gulonate Reductase (YiaK).
J.Biol.Chem., 279, 2004
|
|
5T25
 
 | | Kinetic, Spectral and Structural Characterization of the Slow Binding Inhibitor Acetopyruvate with Dihydrodipicolinate Synthase from Escherichia coli. | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, LYSINE, SODIUM ION | | Authors: | Chooback, L, Thomas, L.M, Karsten, W.E, Fleming, C.D, Seabourn, P. | | Deposit date: | 2016-08-23 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Kinetic, Spectral and Structural Characterization of the Slow Binding Inhibitor Acetopyruvate with Dihydrodipicolinate Synthase from Escherichia coli.
To Be Published
|
|
