4L13
 
 | |
5BRH
 
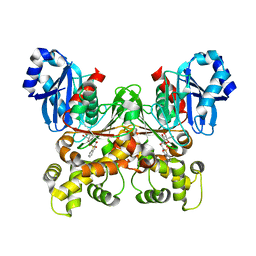 | | Crystal structure of Trypanosoma cruzi glucokinase in complex with inhibitor DBT-GlcN | | Descriptor: | 2-deoxy-2-({[(1,1-dioxido-1-benzothiophen-2-yl)methoxy]carbonyl}amino)-beta-D-glucopyranose, Glucokinase 1, putative | | Authors: | D'Antonio, E.L, Perry, K, Deinema, M.S, Kearns, S.P, Frey, T.A. | | Deposit date: | 2015-05-30 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based approach to the identification of a novel group of selective glucosamine analogue inhibitors of Trypanosoma cruzi glucokinase.
Mol.Biochem.Parasitol., 204, 2016
|
|
5BRF
 
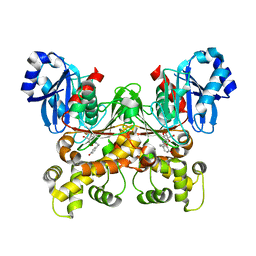 | | Crystal structure of Trypanosoma cruzi glucokinase in complex with inhibitor HPOP-GlcN | | Descriptor: | 2-deoxy-2-{[3-(4-hydroxyphenyl)propanoyl]amino}-alpha-D-glucopyranose, Glucokinase 1, putative | | Authors: | D'Antonio, E.L, Perry, K, Deinema, M.S, Kearns, S.P, Frey, T.A. | | Deposit date: | 2015-05-30 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structure-based approach to the identification of a novel group of selective glucosamine analogue inhibitors of Trypanosoma cruzi glucokinase.
Mol.Biochem.Parasitol., 204, 2016
|
|
4OJ2
 
 | |
5XF5
 
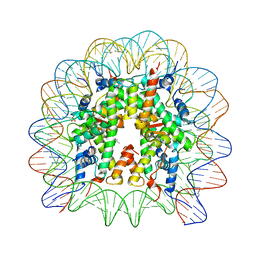 | | Nucleosome core particle with an adduct of a binuclear RAPTA (Ru-arene-phosphaadamantane) compound having a 1,2-diphenylethylenediamine linker (R,S-configuration) | | Descriptor: | (1S,2R)-1,2-diphenylethane-1,2-diamine, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Ma, Z, Adhireksan, Z, Murray, B.S, Dyson, P.J, Davey, C.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Nucleosome acidic patch-targeting binuclear ruthenium compounds induce aberrant chromatin condensation
Nat Commun, 8, 2017
|
|
5XF6
 
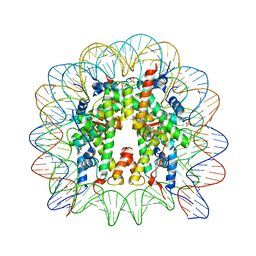 | | Nucleosome core particle with an adduct of a binuclear RAPTA (Ru-arene-phosphaadamantane) compound having an ethylenediamine linker | | Descriptor: | DNA (145-MER), ETHANE-1,2-DIAMINE, Histone H2A, ... | | Authors: | Ma, Z, Adhireksan, Z, Murray, B.S, Dyson, P.J, Davey, C.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Nucleosome acidic patch-targeting binuclear ruthenium compounds induce aberrant chromatin condensation
Nat Commun, 8, 2017
|
|
5YIA
 
 | | Crystal Structure of KNI-10343 bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-[[(2R)-2-[2-(4-hydroxyphenyl)ethanoylamino]-3-methylsulfanyl-propanoyl]amino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-N-[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]-1,3-thiazolidine-4-carboxamide, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, GLYCEROL, ... | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2017-10-03 | | Release date: | 2018-07-11 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Deciphering the mechanism of potent peptidomimetic inhibitors targeting plasmepsins - biochemical and structural insights.
Febs J., 285, 2018
|
|
5YIE
 
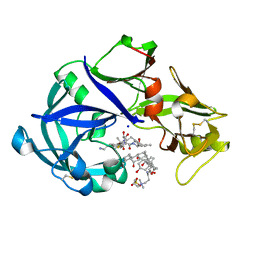 | | Crystal Structure of KNI-10742 bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-[2-[4-[2-azanylethyl(ethyl)amino]-2,6-dimethyl-phenoxy]ethanoylamino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-N-[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]-1,3-thiazolidine-4-carboxamide, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Plasmepsin II, ... | | Authors: | Mishra, V, Rathore, I, Bhaumik, P. | | Deposit date: | 2017-10-04 | | Release date: | 2018-07-11 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Deciphering the mechanism of potent peptidomimetic inhibitors targeting plasmepsins - biochemical and structural insights.
Febs J., 285, 2018
|
|
5YIC
 
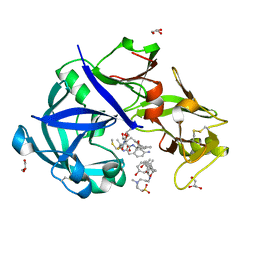 | | Crystal Structure of KNI-10333 bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-[[(2R)-2-[2-(4-aminophenyl)ethanoylamino]-3-methylsulfanyl-propanoyl]amino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-N-[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]-1,3-thiazolidine-4-carboxamide, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, GLYCEROL, ... | | Authors: | Mishra, V, Rathore, I, Bhaumik, P. | | Deposit date: | 2017-10-03 | | Release date: | 2018-07-11 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Deciphering the mechanism of potent peptidomimetic inhibitors targeting plasmepsins - biochemical and structural insights.
Febs J., 285, 2018
|
|
5YIB
 
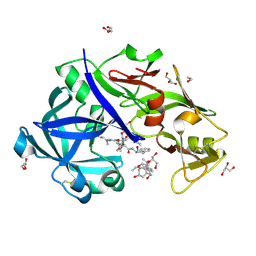 | | Crystal Structure of KNI-10743 bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-[2-[4-[2-(dimethylamino)ethyl-methyl-amino]-2,6-dimethyl-phenoxy]ethanoylamino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-N-[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]-1,3-thiazolidine-4-carboxamide, 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, ... | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2017-10-03 | | Release date: | 2018-07-11 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Deciphering the mechanism of potent peptidomimetic inhibitors targeting plasmepsins - biochemical and structural insights.
Febs J., 285, 2018
|
|
7DJI
 
 | |
4EF3
 
 | | Multicopper Oxidase CueO (Citrate buffer) | | Descriptor: | Blue copper oxidase CueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Komori, H, Kataoka, K, Sakurai, T, Higuchi, Y. | | Deposit date: | 2012-03-29 | | Release date: | 2013-05-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exogenous acetate ion reaches the type II copper centre in CueO through the water-excretion channel and potentially affects the enzymatic activity.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5YID
 
 | | Crystal Structure of KNI-10395 bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | (4R)-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-3-[(2S,3S)-2-hydroxy-3-{[S-methyl-N-(phenylacetyl)-L-cysteinyl]amino}-4-phenylbutanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Plasmepsin II, ... | | Authors: | Mishra, V, Rathore, I, Bhaumik, P. | | Deposit date: | 2017-10-04 | | Release date: | 2018-07-11 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Deciphering the mechanism of potent peptidomimetic inhibitors targeting plasmepsins - biochemical and structural insights.
Febs J., 285, 2018
|
|
4QN0
 
 | | Crystal structure of the CPS-6 mutant Q130K | | Descriptor: | Endonuclease G, mitochondrial, MAGNESIUM ION | | Authors: | Lin, J.L.J, Yuan, H.S. | | Deposit date: | 2014-06-17 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Oxidative Stress Impairs Cell Death by Repressing the Nuclease Activity of Mitochondrial Endonuclease G
Cell Rep, 16, 2016
|
|
5XF4
 
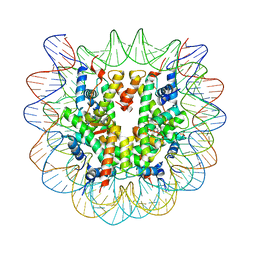 | | Nucleosome core particle with an adduct of a binuclear RAPTA (Ru-arene-phosphaadamantane) compound having a 1,2-diphenylethylenediamine linker (S,S-configuration) | | Descriptor: | (1S,2S)-1,2-diphenylethane-1,2-diamine, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Ma, Z, Adhireksan, Z, Murray, B.S, Dyson, P.J, Davey, C.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Nucleosome acidic patch-targeting binuclear ruthenium compounds induce aberrant chromatin condensation
Nat Commun, 8, 2017
|
|
5ZLQ
 
 | | Crystal Structure of C1orf123 | | Descriptor: | UPF0587 protein C1orf123, ZINC ION | | Authors: | Furukawa, Y, Lim, C.T, Tosha, T. | | Deposit date: | 2018-03-29 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a novel zinc-binding protein, C1orf123, as an interactor with a heavy metal-associated domain
PLoS ONE, 13, 2018
|
|
5YY9
 
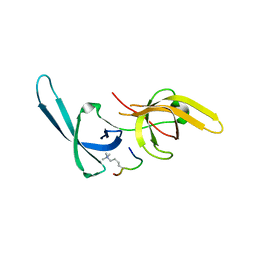 | | Crystal structure of Tandem Tudor Domain of human UHRF1 in complex with LIG1-K126me3 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Ligase 1 | | Authors: | Kori, S, Defossez, P.A, Arita, K. | | Deposit date: | 2017-12-08 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Structure of the UHRF1 Tandem Tudor Domain Bound to a Methylated Non-histone Protein, LIG1, Reveals Rules for Binding and Regulation.
Structure, 27, 2019
|
|
7K3P
 
 | |
5YYA
 
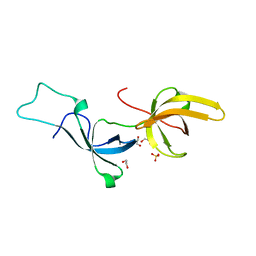 | | Crystal structure of Tandem Tudor Domain of human UHRF1 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1, SULFATE ION | | Authors: | Kori, S, Defossez, P.A, Arita, K. | | Deposit date: | 2017-12-08 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the UHRF1 Tandem Tudor Domain Bound to a Methylated Non-histone Protein, LIG1, Reveals Rules for Binding and Regulation.
Structure, 27, 2019
|
|
5XF3
 
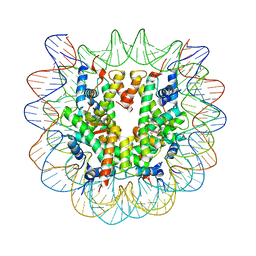 | | Nucleosome core particle with an adduct of a binuclear RAPTA (Ru-arene-phosphaadamantane) compound having a 1,2-diphenylethylenediamine linker (R,R-configuration) | | Descriptor: | (1R,2R)-1,2-diphenylethane-1,2-diamine, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Ma, Z, Adhireksan, Z, Murray, B.S, Dyson, P.J, Davey, C.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nucleosome acidic patch-targeting binuclear ruthenium compounds induce aberrant chromatin condensation
Nat Commun, 8, 2017
|
|
1NDN
 
 | | MOLECULAR STRUCTURE OF NICKED DNA. MODEL T4 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*AP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*T)-3'), DNA (5'-D(*TP*TP*CP*GP*CP*G)-3') | | Authors: | Aymani, J, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1992-01-15 | | Release date: | 1992-07-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular structure of nicked DNA: a substrate for DNA repair enzymes.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
7VH9
 
 | | Solution structure of the chimeric peptide of the first SURP domain of Human SF3A1 and the interacting region of SF1. | | Descriptor: | Splicing factor 3A subunit 1,Splicing factor 1 | | Authors: | Muto, Y, Kuwasako, K, Takizawa, M, Kobayashi, N, Sakamoto, T. | | Deposit date: | 2021-09-21 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the interaction between the first SURP domain of the SF3A1 subunit in U2 snRNP and the human splicing factor SF1.
Protein Sci., 31, 2022
|
|
5BNA
 
 | | THE PRIMARY MODE OF BINDING OF CISPLATIN TO A B-DNA DODECAMER: C-G-C-G-A-A-T-T-C-G-C-G | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PLATINUM TRIAMINE ION | | Authors: | Wing, R.M, Pjura, P, Drew, H.R, Dickerson, R.E. | | Deposit date: | 1983-08-22 | | Release date: | 1983-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The primary mode of binding of cisplatin to a B-DNA dodecamer: C-G-C-G-A-A-T-T-C-G-C-G
EMBO J., 3, 1984
|
|
7BD4
 
 | | Notum Fragment 828 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-(4-methoxyphenyl)-N'-pyridin-4-ylurea, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BDD
 
 | | Notum Fragment 924 | | Descriptor: | (2~{R})-2-(4-phenylphenoxy)propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
