8XQL
 
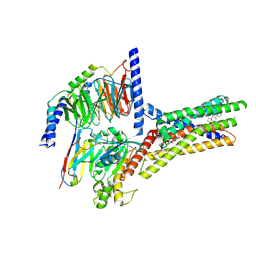 | | Structure of human class T GPCR TAS2R14-miniGs/gust complex with Aristolochic acid A. | | Descriptor: | 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
1VSC
 
 | | VCAM-1 | | Descriptor: | VASCULAR CELL ADHESION MOLECULE-1 | | Authors: | Wang, J, Stehle, T, Osborn, L. | | Deposit date: | 1995-04-27 | | Release date: | 1996-06-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of an N-terminal two-domain fragment of vascular cell adhesion molecule 1 (VCAM-1): a cyclic peptide based on the domain 1 C-D loop can inhibit VCAM-1-alpha 4 integrin interaction.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
6XY2
 
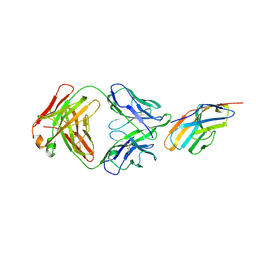 | | Crystal structure of CTLA-4 complexed with the Fab of HL32 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytotoxic T-lymphocyte protein 4, Heavy chain, ... | | Authors: | Gao, H, Zhou, A. | | Deposit date: | 2020-01-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of CTLA-4 complexed with a pH-sensitive cancer immunotherapeutic antibody.
Cell Discov, 6, 2020
|
|
1LSL
 
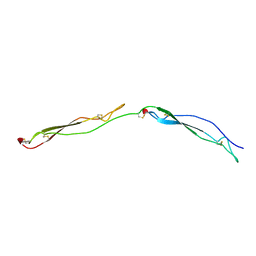 | | Crystal Structure of the Thrombospondin-1 Type 1 Repeats | | Descriptor: | Thrombospondin 1, alpha-L-fucopyranose, beta-L-fucopyranose | | Authors: | Tan, K, Duquette, M, Liu, J, Dong, Y, Zhang, R, Joachimiak, A, Lawler, J, Wang, J.-H. | | Deposit date: | 2002-05-17 | | Release date: | 2002-12-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the TSP-1 type 1 repeats: a novel
layered fold and its biological implication.
J.Cell Biol., 159, 2002
|
|
3H3B
 
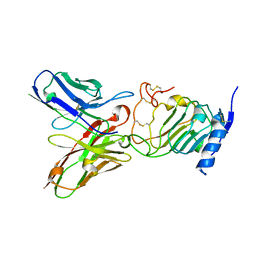 | | Crystal structure of the single-chain Fv (scFv) fragment of an anti-ErbB2 antibody chA21 in complex with residues 1-192 of ErbB2 extracellular domain | | Descriptor: | Receptor tyrosine-protein kinase erbB-2, anti-ErbB2 antibody chA21 | | Authors: | Zhou, H, Liu, Y, Niu, L, Zhu, J, Teng, M. | | Deposit date: | 2009-04-16 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Insights into the Down-regulation of Overexpressed p185her2/neu Protein of Transformed Cells by the Antibody chA21.
J.Biol.Chem., 286, 2011
|
|
7FCP
 
 | |
7FCQ
 
 | |
8AYR
 
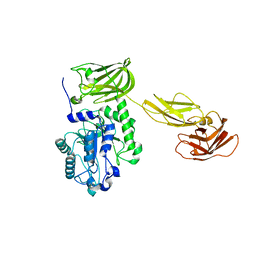 | | Sialidases and Fucosidases of Akkermansia muciniphila are key for rapid growth on colonic mucin and nutrient sharing amongst mucin-associated human gut microbiota | | Descriptor: | CALCIUM ION, Coagulation factor 5/8 type domain protein | | Authors: | Sakanaka, H, Nielsen, T.S, Pichler, M.J, Nordberg Karlsson, E, Abou Hachem, M, Morth, J.P. | | Deposit date: | 2022-09-02 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sialidases and fucosidases of Akkermansia muciniphila are crucial for growth on mucin and nutrient sharing with mucus-associated gut bacteria.
Nat Commun, 14, 2023
|
|
8AXT
 
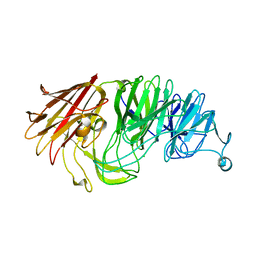 | | Sialidases and Fucosidases of Akkermansia muciniphila are key for rapid growth on colonic mucin and nutrient sharing amongst mucin-associated human gut microbiota | | Descriptor: | CALCIUM ION, CHLORIDE ION, Sialidase domain-containing protein | | Authors: | Sakanaka, H, Nielsen, T.S, Pichler, M.J, Nordberg Karlsson, E, Abou Hachem, M, Morth, J.P. | | Deposit date: | 2022-08-31 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Sialidases and fucosidases of Akkermansia muciniphila are crucial for growth on mucin and nutrient sharing with mucus-associated gut bacteria.
Nat Commun, 14, 2023
|
|
8AXS
 
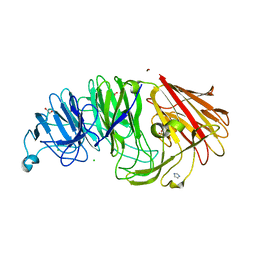 | | Sialidases and Fucosidases of Akkermansia muciniphila are key for rapid growth on colonic mucin and nutrient sharing amongst mucin-associated human gut microbiota | | Descriptor: | 1,2-ETHANEDIOL, 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, ... | | Authors: | Sakanaka, H, Nielsen, T.S, Pichler, M.J, Nordberg Karlsson, E, Abou Hachem, M, Morth, J.P. | | Deposit date: | 2022-08-31 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Sialidases and fucosidases of Akkermansia muciniphila are crucial for growth on mucin and nutrient sharing with mucus-associated gut bacteria.
Nat Commun, 14, 2023
|
|
8AXI
 
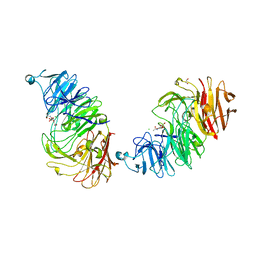 | | Sialidases and Fucosidases of Akkermansia muciniphila are key for rapid growth on colonic mucin and nutrient sharing amongst mucin-associated human gut microbiota | | Descriptor: | 1,2-ETHANEDIOL, 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, ... | | Authors: | Sakanaka, H, Nielsen, T.S, Pichler, M.J, Nordberg Karlsson, E, Abou Hachem, M, Morth, J.P. | | Deposit date: | 2022-08-31 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Sialidases and fucosidases of Akkermansia muciniphila are crucial for growth on mucin and nutrient sharing with mucus-associated gut bacteria.
Nat Commun, 14, 2023
|
|
6WBW
 
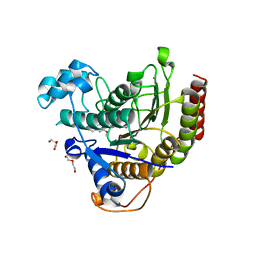 | | Structure of Human HDAC2 in complex with an ethyl ketone inhibitor | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Histone deacetylase 2, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2020-03-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of ethyl ketone-based HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6WBZ
 
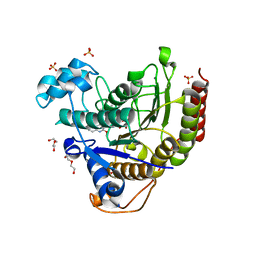 | | Structure of Human HDAC2 in complex with an ethyl ketone inhibitor containing a spiro-bicyclic group | | Descriptor: | (1S)-N-{(1S)-7,7-dihydroxy-1-[5-(2-methoxyquinolin-3-yl)-1H-imidazol-2-yl]nonyl}-6-ethyl-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2020-03-28 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Discovery of ethyl ketone-based HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
3J1P
 
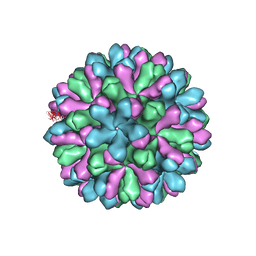 | | Atomic model of rabbit hemorrhagic disease virus | | Descriptor: | Major capsid protein VP60 | | Authors: | Wang, X, Liu, Y, Sun, F. | | Deposit date: | 2012-04-09 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography.
Plos Pathog., 9, 2013
|
|
1D9K
 
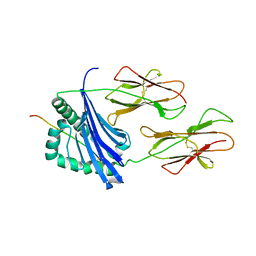 | | CRYSTAL STRUCTURE OF COMPLEX BETWEEN D10 TCR AND PMHC I-AK/CA | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CONALBUMIN PEPTIDE, ... | | Authors: | Reinherz, E.L, Tan, K, Tang, L, Kern, P, Liu, J.-H, Xiong, Y, Hussey, R.E, Smolyar, A, Hare, B, Zhang, R, Joachimiak, A, Chang, H.-C, Wagner, G, Wang, J.-H. | | Deposit date: | 1999-10-28 | | Release date: | 1999-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of a T cell receptor in complex with peptide and MHC class II.
Science, 286, 1999
|
|
4M57
 
 | | Crystal structure of the pentatricopeptide repeat protein PPR10 from maize | | Descriptor: | Chloroplast pentatricopeptide repeat protein 10 | | Authors: | Yin, P, Li, Q, Yan, C, Liu, Y, Yan, N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for the modular recognition of single-stranded RNA by PPR proteins.
Nature, 504, 2013
|
|
5GTY
 
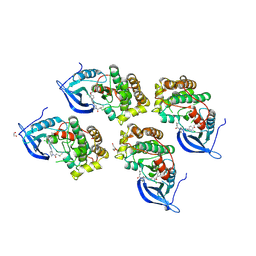 | | Crystal structure of EGFR 696-1022 T790M in complex with LXX-6-26 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(3R)-3-[4-azanyl-3-[3-chloranyl-4-[(6-methylpyridin-2-yl)methoxy]phenyl]pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]prop-2-en-1-one, Epidermal growth factor receptor | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Discovery and characterization of a novel irreversible EGFR mutants selective and potent kinase inhibitor CHMFL-EGFR-26 with a distinct binding mode.
Oncotarget, 8, 2017
|
|
3EVX
 
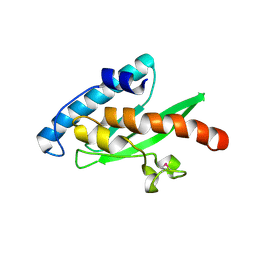 | | Crystal structure of the human E2-like ubiquitin-fold modifier conjugating enzyme 1 (Ufc1). Northeast Structural Genomics Consortium target HR41 | | Descriptor: | THIOCYANATE ION, Ufm1-conjugating enzyme 1 | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Ho, C.K, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-13 | | Release date: | 2008-10-21 | | Last modified: | 2023-01-04 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | NMR and X-RAY structures of human E2-like ubiquitin-fold modifier conjugating enzyme 1 (UFC1) reveal structural and functional conservation in the metazoan UFM1-UBA5-UFC1 ubiquination pathway.
J.STRUCT.FUNCT.GENOM., 10, 2009
|
|
2QKW
 
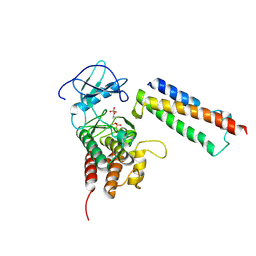 | | Structural basis for activation of plant immunity by bacterial effector protein AvrPto | | Descriptor: | Avirulence protein, Protein kinase | | Authors: | Xing, W.M, Zou, Y, Liu, Q, Hao, Q, Zhou, J.M, Chai, J.J. | | Deposit date: | 2007-07-11 | | Release date: | 2007-08-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis for activation of plant immunity by bacterial effector protein AvrPto
Nature, 449, 2007
|
|
6XEC
 
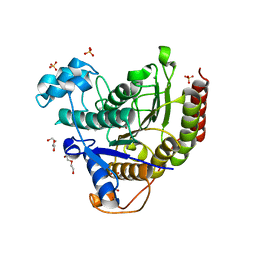 | | STRUCTURE OF HUMAN HDAC2 IN COMPLEX WITH KETONE INHIBITOR (COMPOUND O) | | Descriptor: | (1S)-N-{(1S)-1-[5-cyano-2-(4-fluorophenyl)-1H-imidazol-4-yl]-7,7-dihydroxynonyl}-6-methyl-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Clausen, D. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Development of a selective HDAC inhibitor aimed at reactivating the HIV latent reservoir.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
8EX1
 
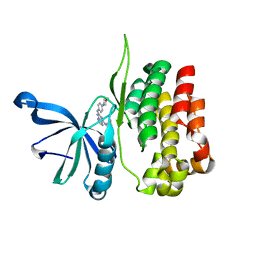 | |
8EX0
 
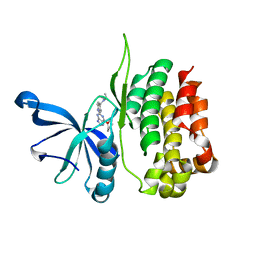 | |
8EX2
 
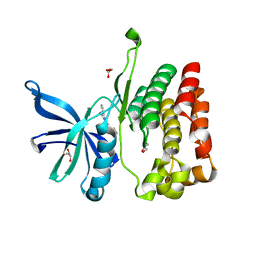 | |
6XEB
 
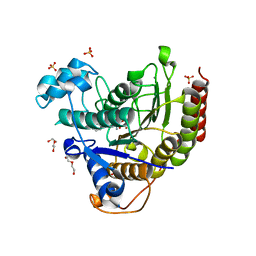 | | STRUCTURE OF HUMAN HDAC2 IN COMPLEX WITH KETONE INHIBITOR (COMPOUND E) | | Descriptor: | 5-{(1S)-7,7-dihydroxy-1-[(1-methylazetidine-3-carbonyl)amino]nonyl}-2-phenyl-1H-imidazole-4-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Clausen, D. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Development of a selective HDAC inhibitor aimed at reactivating the HIV latent reservoir.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
7QUR
 
 | | SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C3 symmetry | | Descriptor: | 2,3,5-tris(iodanyl)benzamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Naismith, J.H, Yang, Y, Liu, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-01 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
