7UV5
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S/D286N mutant, in complex with a Lys48-linked di-ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, Papain-like protease nsp3, Ubiquitin, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-04-29 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
4ZWL
 
 | | 2.60 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) H448F/Y450L double mutant from Staphylococcus aureus in complex with NAD+ and BME-free Cys289 | | Descriptor: | Betaine-aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-19 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.60 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) H448F/Y450L double mutant from Staphylococcus aureus in complex with NAD+ and BME-free Cys289
To be Published
|
|
5JG7
 
 | | Crystal structure of putative periplasmic binding protein from Salmonella typhimurium LT2 | | Descriptor: | Fur regulated Salmonella iron transporter, GLYCEROL | | Authors: | Chang, C, Zhou, M, Shatsman, S, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-04-19 | | Release date: | 2016-04-27 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative periplasmic binding protein from Salmonella typhimurium LT2
To Be Published
|
|
6VEK
 
 | | Contact-dependent growth inhibition toxin-immunity protein complex from from E. coli 3006, full-length | | Descriptor: | contact-dependent immunity protein CdiI, contact-dependent toxin CdiA | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-02 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Contact-dependent growth inhibition toxin-immunity protein complex from from E. coli 3006, full-length
To Be Published
|
|
6DUX
 
 | | 2.25 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Klebsiella pneumoniae in Complex with NAD. | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 6-phospho-alpha-glucosidase, ACETATE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-22 | | Release date: | 2018-07-04 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
4NOG
 
 | | Crystal structure of a putative ornithine aminotransferase from Toxoplasma gondii ME49 in complex with pyrodoxal-5'-phosphate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Filippova, E.V, Halavaty, A, Ruan, J, Shuvalova, L, Flores, K, Dubrovska, I, Ngo, H, Shanmugam, D, Roos, D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
5KL9
 
 | | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with CoA | | Descriptor: | Acyl-CoA thioester hydrolase YbgC, COENZYME A, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-23 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with CoA
To Be Published
|
|
5JQ4
 
 | | Structure of a GNAT acetyltransferase SACOL1063 from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase SACOL1063, CHLORIDE ION, ... | | Authors: | Majorek, K.A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-04 | | Release date: | 2016-06-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insight into the 3D structure and substrate specificity of previously uncharacterized GNAT superfamily acetyltransferases from pathogenic bacteria.
Biochim.Biophys.Acta, 1865, 2016
|
|
6VWW
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2. | | Descriptor: | ACETIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-20 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Protein Sci., 29, 2020
|
|
6DVV
 
 | | 2.25 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Klebsiella pneumoniae in Complex with NAD and Mn2+. | | Descriptor: | 6-phospho-alpha-glucosidase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-25 | | Release date: | 2018-07-18 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6W01
 
 | | The 1.9 A Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with a Citrate | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-28 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Protein Sci., 29, 2020
|
|
4OHN
 
 | | Crystal structure of an ABC uptake transporter substrate binding protein from Streptococcus pneumoniae with Bound Histidine | | Descriptor: | ABC transporter substrate-binding protein, ACETATE ION, HISTIDINE | | Authors: | Brunzelle, J.S, Wawrzak, W, Yim, Y, Kudritska, M, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of an ABC uptake transporter substrate binding protein from Streptococcus pneumoniae with Bound Histidine
To be Published
|
|
6DT4
 
 | | 1.8 Angstrom Resolution Crystal Structure of cAMP-Regulatory Protein from Yersinia pestis in Complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CHLORIDE ION, Cyclic AMP receptor protein | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Ritzert, J.T.H, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-15 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Cyclic AMP Receptor Protein Regulates Quorum Sensing and Global Gene Expression in Yersinia pestis during Planktonic Growth and Growth in Biofilms.
Mbio, 10, 2019
|
|
5F48
 
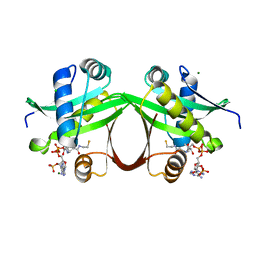 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample in complex with coenzyme A | | Descriptor: | CHLORIDE ION, COENZYME A, MAGNESIUM ION, ... | | Authors: | Xu, Z, Skarina, T, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
6E9P
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - open form with BRD0059 bound | | Descriptor: | (2R,3S,4R)-3-(2',6'-difluoro-4'-methyl[1,1'-biphenyl]-4-yl)-4-(fluoromethyl)azetidine-2-carbonitrile, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-08-01 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.569 Å) | | Cite: | Crystal structure of tryptophan synthase from M. tuberculosis - closed form with BRD6309 bound
To be Published
|
|
5VM2
 
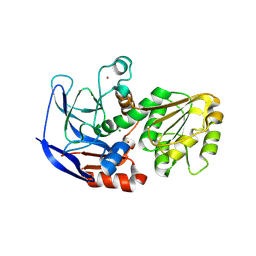 | | Crystal structure of ECK1772, an oxidoreductase/dehydrogenase of unknown specificity involved in membrane biogenesis from Escherichia coli | | Descriptor: | Alcohol dehydrogenase, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Stogios, P.J, Skarina, T, McChesney, C, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-26 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Crystal structure of ECK1772, an oxidoreductase/dehydrogenase of unknown specificity involved in membrane biogenesis from Escherichia coli
To Be Published
|
|
3OF5
 
 | | Crystal Structure of a Dethiobiotin Synthetase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | ACETATE ION, Dethiobiotin synthetase, SODIUM ION | | Authors: | Brunzelle, J.S, Skarina, T, Gordon, E, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-13 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of a Dethiobiotin Synthetase from Francisella tularensis subsp. tularensis SCHU S4
TO BE PUBLISHED
|
|
6P7U
 
 | |
5F49
 
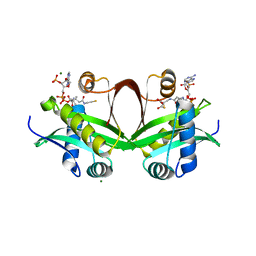 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample in complex with malonyl-coenzyme A | | Descriptor: | COENZYME A, MAGNESIUM ION, MALONYL-COENZYME A, ... | | Authors: | Xu, Z, Skarina, T, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
4YFJ
 
 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-Ib | | Descriptor: | Aminoglycoside 3'-N-acetyltransferase, SULFATE ION | | Authors: | Stogios, P.J, Xu, Z, Evdokimova, E, Yim, V, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of aminoglycoside acetyltransferase AAC(3)-Ib
To Be Published
|
|
5WP0
 
 | | Crystal structure of NAD synthetase NadE from Vibrio fischeri | | Descriptor: | NH(3)-dependent NAD(+) synthetase | | Authors: | Stogios, P.J, Evdokimova, E, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-08-03 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of NAD synthetase NadE from Vibrio fischeri
To Be Published
|
|
7JM1
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, Aminocyclitol acetyltransferase ApmA | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-30 | | Release date: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with acetyl-CoA
To Be Published
|
|
7KAG
 
 | | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 3, SULFATE ION | | Authors: | Stogios, P.J, Skarina, T, Chang, C, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-30 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2
To Be Published
|
|
5VYV
 
 | | Crystal structure of a protein of unknown function YceH/ECK1052 involved in membrane biogenesis from Escherichia coli | | Descriptor: | DI(HYDROXYETHYL)ETHER, UPF0502 protein YceH | | Authors: | Stogios, P.J, Skarina, T, McChesney, C, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-05-26 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of a protein of unknown function YceH/ECK1052 involved in membrane biogenesis from Escherichia coli
To Be Published
|
|
4YPI
 
 | | Structure of Ebola virus nucleoprotein N-terminal fragment bound to a peptide derived from Ebola VP35 | | Descriptor: | Nucleoprotein, Polymerase cofactor VP35 | | Authors: | Leung, D.W, Borek, D.M, Binning, J.M, Otwinowski, Z, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-03-13 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | An Intrinsically Disordered Peptide from Ebola Virus VP35 Controls Viral RNA Synthesis by Modulating Nucleoprotein-RNA Interactions.
Cell Rep, 11, 2015
|
|
