6WAA
 
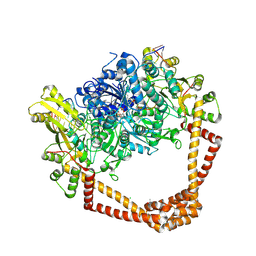 | | K. pneumoniae Topoisomerase IV (ParE-ParC) in complex with DNA and compound 34 (7-[(1S,5R)-1-amino-3-azabicyclo[3.1.0]hexan-3-yl]-4-(aminomethyl)-1-cyclopropyl-3,6-difluoro-8-methylquinolin-2(1H)-one) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-[(1S,5R)-1-amino-3-azabicyclo[3.1.0]hexan-3-yl]-4-(aminomethyl)-1-cyclopropyl-3,6-difluoro-8-methylquinolin-2(1H)-one, CHLORIDE ION, ... | | Authors: | Noeske, J, Shu, W, Bellamacina, C. | | Deposit date: | 2020-03-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Topoisomerase Inhibitors Addressing Fluoroquinolone Resistance in Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
6LEB
 
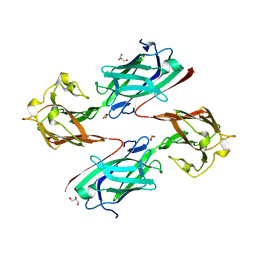 | | Staphylococcus aureus surface protein SdrC mutant-P366H | | Descriptor: | GLYCEROL, MAGNESIUM ION, Ser-Asp rich fibrinogen-binding, ... | | Authors: | Hang, T, Zhang, M, Wang, J. | | Deposit date: | 2019-11-25 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural insights into the intermolecular interaction of the adhesin SdrC in the pathogenicity of Staphylococcus aureus.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
8WYP
 
 | |
6KDV
 
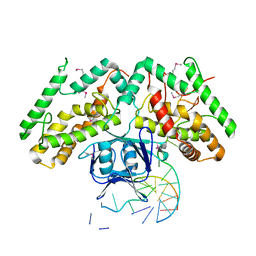 | | Crystal structure of TtCas1-DNA complex | | Descriptor: | CRISPR-associated endonuclease Cas1 2, DNA (5'-D(*GP*AP*GP*TP*CP*GP*AP*TP*GP*CP*TP*GP*GP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*CP*CP*AP*GP*CP*AP*TP*CP*GP*AP*CP*TP*C)-3') | | Authors: | Wang, Y.L, Li, J.Z, Yang, J, Wang, J.Y. | | Deposit date: | 2019-07-02 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure of Cas1 in complex with branched DNA.
Sci China Life Sci, 63, 2020
|
|
8HM2
 
 | |
8HM1
 
 | |
6KE1
 
 | | Crystal structure of TtCas1 | | Descriptor: | CRISPR-associated endonuclease Cas1 2 | | Authors: | Wang, Y.L, Li, J.Z, Yang, J, Wang, J.Y. | | Deposit date: | 2019-07-03 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Crystal structure of Cas1 in complex with branched DNA.
Sci China Life Sci, 63, 2020
|
|
4IJV
 
 | | Crystal structure of 11b-HSD1 double mutant (L262R, F278E) in complex with 3-[1-(4-chlorophenyl)cyclopropyl]-8-(2-fluorophenoxy)[1,2,4]triazolo[4,3-a]pyridine | | Descriptor: | 3-[1-(4-chlorophenyl)cyclopropyl]-8-(2-fluorophenoxy)[1,2,4]triazolo[4,3-a]pyridine, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2012-12-23 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Optimization of 1,2,4-Triazolopyridines as Inhibitors of Human 11 beta-Hydroxysteroid Dehydrogenase Type 1 (11 beta-HSD-1).
ACS Med Chem Lett, 5, 2014
|
|
3SPZ
 
 | | DNA Polymerase(L415A/L561A/S565G/Y567A) Ternary Complex with dUpCpp Opposite dA (Ca2+) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]uridine, 5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*C)-3', 5'-D(P*GP*AP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3', ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-04 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural Insights into Complete Metal Ion Coordination from Ternary Complexes of B Family RB69 DNA Polymerase.
Biochemistry, 50, 2011
|
|
4IJW
 
 | | Crystal structure of 11b-HSD1 double mutant (L262R, F278E) in complex with 3-[1-(4-chlorophenyl)cyclopropyl]-8-cyclopropyl[1,2,4]triazolo[4,3-a]pyridine | | Descriptor: | 3-[1-(4-chlorophenyl)cyclopropyl]-8-cyclopropyl[1,2,4]triazolo[4,3-a]pyridine, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2012-12-23 | | Release date: | 2014-06-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Optimization of 1,2,4-Triazolopyridines as Inhibitors of Human 11 beta-Hydroxysteroid Dehydrogenase Type 1 (11 beta-HSD-1).
ACS Med Chem Lett, 5, 2014
|
|
2QDY
 
 | | Crystal Structure of Fe-type NHase from Rhodococcus erythropolis AJ270 | | Descriptor: | 2-METHYLPROPAN-1-AMINE, CHLORIDE ION, FE (III) ION, ... | | Authors: | Song, L, Shi, J, Xue, Z, Wang, M.-X, Qian, S. | | Deposit date: | 2007-06-22 | | Release date: | 2008-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High resolution X-ray molecular structure of the nitrile hydratase from Rhodococcus erythropolis AJ270 reveals posttranslational oxidation of two cysteines into sulfinic acids and a novel biocatalytic nitrile hydration mechanism
Biochem.Biophys.Res.Commun., 362, 2007
|
|
3BU7
 
 | | Crystal Structure and Biochemical Characterization of GDOsp, a Gentisate 1,2-Dioxygenase from Silicibacter Pomeroyi | | Descriptor: | FE (II) ION, Gentisate 1,2-dioxygenase | | Authors: | Chen, J, Wang, M.Z, Zhu, G.Y, Zhang, X.C, Rao, Z.H. | | Deposit date: | 2008-01-02 | | Release date: | 2008-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and mutagenic analysis of GDOsp, a gentisate 1,2-dioxygenase from Silicibacter pomeroyi.
Protein Sci., 17, 2008
|
|
6IMM
 
 | | Cryo-EM structure of an alphavirus, Sindbis virus | | Descriptor: | Assembly protein E3, Octadecane, Spike glycoprotein E1, ... | | Authors: | Zhang, X, Ma, J, Chen, L. | | Deposit date: | 2018-10-23 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Implication for alphavirus host-cell entry and assembly indicated by a 3.5 angstrom resolution cryo-EM structure.
Nat Commun, 9, 2018
|
|
6IFL
 
 | | Cryo-EM structure of type III-A Csm-NTR complex | | Descriptor: | NTR, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-20 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
8WZ8
 
 | | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021(T67A,Q69A) complex with NAD | | Descriptor: | Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Gao, Y.S, Xie, R, Chen, Y.N, Ma, J.M, Ge, H.H. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.583 Å) | | Cite: | Structural basis for substrate recognition by a S-adenosylhomocysteine hydrolase Lpg2021 from Legionella pneumophila.
Int.J.Biol.Macromol., 270, 2024
|
|
8WZ6
 
 | | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021 in ternary complex with NAD and DZNep | | Descriptor: | Adenosylhomocysteinase, DZNep, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Gao, Y.S, Xie, R, Chen, Y.N, Ma, J.M, Ge, H.H. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural basis for substrate recognition by a S-adenosylhomocysteine hydrolase Lpg2021 from Legionella pneumophila.
Int.J.Biol.Macromol., 270, 2024
|
|
8WZ7
 
 | | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021(I255A,T287A) in ternary complex with NAD and adenosine | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Gao, Y.S, Xie, R, Chen, Y.N, Ma, J.M, Ge, H.H. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Structural basis for substrate recognition by a S-adenosylhomocysteine hydrolase Lpg2021 from Legionella pneumophila.
Int.J.Biol.Macromol., 270, 2024
|
|
8WWG
 
 | |
8WZ9
 
 | | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021 in ternary complex with NAD and adenine | | Descriptor: | ADENINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Gao, Y.S, Xie, R, Chen, Y.N, Ma, J.M, Ge, H.H. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Structural basis for substrate recognition by a S-adenosylhomocysteine hydrolase Lpg2021 from Legionella pneumophila.
Int.J.Biol.Macromol., 270, 2024
|
|
3E9U
 
 | | Crystal structure of Calx CBD2 domain | | Descriptor: | Na/Ca exchange protein | | Authors: | Wu, M, Zheng, L. | | Deposit date: | 2008-08-23 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CBD2 from the Drosophila Na(+)/Ca(2+) exchanger: diversity of Ca(2+) regulation and its alternative splicing modification.
J.Mol.Biol., 387, 2009
|
|
2W42
 
 | | THE STRUCTURE OF A PIWI PROTEIN FROM ARCHAEOGLOBUS FULGIDUS COMPLEXED WITH A 16NT DNA DUPLEX. | | Descriptor: | 5'-D(*GP*TP*CP*GP*AP*AP*TP*TP)-3', 5'-D(*TP*TP*CP*GP*AP*CP*GP*CP)-3', MANGANESE (II) ION, ... | | Authors: | Parker, J.S, Roe, S.M, Barford, D. | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Enhancement of the Seed-Target Recognition Step in RNA Silencing by a Piwi-Mid Domain Protein
Mol.Cell, 33, 2009
|
|
5XKI
 
 | |
8S86
 
 | | human PLD3 homodimer structure | | Descriptor: | 5'-3' exonuclease PLD3 | | Authors: | Lammens, K. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Lysosomal endonuclease RNase T2 and PLD exonucleases cooperatively generate RNA ligands for TLR7 activation.
Immunity, 57, 2024
|
|
9JN8
 
 | |
5XTO
 
 | |
