6TCG
 
 | |
6T5S
 
 | | Apo form of C-type lysozyme from the upper gastrointestinal tract of Opisthocomus hoatzin | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C | | Authors: | Taylor, E.J, Skjot, M, Skov, L.K, Klausen, M, De Maria, L, Gippert, G.P, Turkenburg, J.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-10-17 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The C-Type Lysozyme from the upper Gastrointestinal Tract of Opisthocomus hoatzin, the Stinkbird.
Int J Mol Sci, 20, 2019
|
|
6CYN
 
 | | CTX-M-14 N106S/D240G mutant | | Descriptor: | Beta-lactamase | | Authors: | Patel, M.P, Hu, L, Sankaran, B, Brown, C, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2018-04-06 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Synergistic effects of functionally distinct substitutions in beta-lactamase variants shed light on the evolution of bacterial drug resistance.
J. Biol. Chem., 293, 2018
|
|
6TKS
 
 | | Tankyrase 2 in complex with an inhibitor (OM-1720) | | Descriptor: | Tankyrase-2, ZINC ION, ~{N}-[3-[5-(5-ethoxypyridin-2-yl)-4-(2-fluorophenyl)-1,2,4-triazol-3-yl]cyclobutyl]-1,6-naphthyridine-2-carboxamide | | Authors: | Sowa, S.T, Lehtio, L. | | Deposit date: | 2019-11-28 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Preclinical Lead Optimization of a 1,2,4-Triazole Based Tankyrase Inhibitor.
J.Med.Chem., 63, 2020
|
|
6T7Y
 
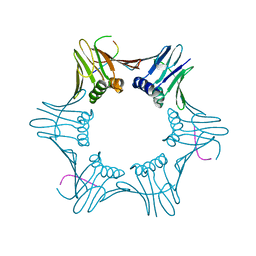 | | Structure of PCNA bound to cPIP motif of DP2 from P. abyssi | | Descriptor: | DNA polymerase sliding clamp, cPIP motif from the DP2 large subunit of PolD | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
6D0J
 
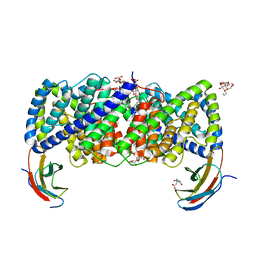 | | Crystal structure of a CLC-type fluoride/proton antiporter | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, CLC-type fluoride/proton antiporter, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Last, N.B, Stockbridge, R.B, Wilson, A.E, Shane, T, Kolmakova-Partensky, L, Koide, A, Koide, S, Miller, C. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A CLC-type F-/H+antiporter in ion-swapped conformations.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6CP8
 
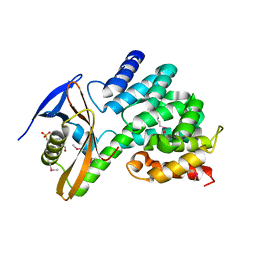 | | Contact-dependent growth inhibition toxin-immunity protein complex from from E. coli 3006 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CdiA, CdiI, ... | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Convergent Evolution of the Barnase/EndoU/Colicin/RelE (BECR) Fold in Antibacterial tRNase Toxins.
Structure, 27, 2019
|
|
6CSB
 
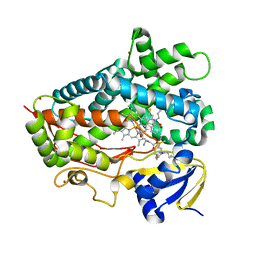 | | V308E mutant of cytochrome P450 2D6 complexed with thioridazine | | Descriptor: | 10-{2-[(2R)-1-methylpiperidin-2-yl]ethyl}-2-(methylsulfanyl)-10H-phenothiazine, ACETATE ION, Cytochrome P450 2D6, ... | | Authors: | Yang, Y.T, Fujita, K, Wang, P.F, Im, S.C, Pearl, N.M, Meagher, J, Stuckey, J, Waskell, L. | | Deposit date: | 2018-03-20 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Characteristic conformational changes on the distal and proximal surfaces of cytochrome P450 2D6 in response to substrate binding
To Be Published
|
|
6CYA
 
 | | Rotavirus SA11 NSP2 S313A mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, Non-structural protein 2 | | Authors: | Anish, R, Hu, L, Prasad, B.V.V. | | Deposit date: | 2018-04-05 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phosphorylation cascade regulates the formation and maturation of rotaviral replication factories.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CYK
 
 | | CTX-M-14 N106S mutant | | Descriptor: | ACETATE ION, Beta-lactamase | | Authors: | Patel, M.P, Hu, L, Sankaran, B, Brown, C, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2018-04-06 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synergistic effects of functionally distinct substitutions in beta-lactamase variants shed light on the evolution of bacterial drug resistance.
J. Biol. Chem., 293, 2018
|
|
6TF2
 
 | |
6D27
 
 | | Crystal structure of the prostaglandin D2 receptor CRTH2 with CAY10471 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, OLEIC ACID, ... | | Authors: | Wang, L, Yao, D, Deepak, K, Liu, H, Gong, W, Fan, H, Wei, Z, Zhang, C. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.738 Å) | | Cite: | Structures of the Human PGD2Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition.
Mol. Cell, 72, 2018
|
|
6D0T
 
 | | De novo design of a fluorescence-activating beta barrel - BB1 | | Descriptor: | BB1 | | Authors: | Dou, J, Vorobieva, A.A, Sheffler, W, Doyle, L.A, Park, H, Bick, M.J, Mao, B, Foight, G.W, Lee, M, Carter, L, Sankaran, B, Ovchinnikov, S, Marcos, E, Huang, P, Vaughan, J.C, Stoddard, B.L, Baker, D. | | Deposit date: | 2018-04-10 | | Release date: | 2018-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
6D1R
 
 | | Structure of Staphylococcus aureus RNase P protein at 2.0 angstrom | | Descriptor: | Ribonuclease P protein component | | Authors: | Ha, L, Colquhoun, J, Noinaj, N, Das, C, Dunman, P, Flaherty, D.P. | | Deposit date: | 2018-04-12 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Crystal structure of the ribonuclease-P-protein subunit from Staphylococcus aureus.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6TG4
 
 | | Tankyrase 2 in complex with an inhibitor (OM-1700) | | Descriptor: | N-[3-[5-(5-ethoxypyridin-2-yl)-4-(2-fluorophenyl)-1,2,4-triazol-3-yl]cyclobutyl]pyridine-2-carboxamide, Tankyrase-2, ZINC ION | | Authors: | Sowa, S.T, Lehtio, L. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Preclinical Lead Optimization of a 1,2,4-Triazole Based Tankyrase Inhibitor.
J.Med.Chem., 63, 2020
|
|
6CYU
 
 | | Crystal structure of CTX-M-14 S70G/N106S/D240G beta-lactamase in complex with hydrolyzed cefotaxime | | Descriptor: | (2R)-2-[(R)-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}(carboxy)methyl]-5-methylidene-5,6-dihydro -2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase | | Authors: | Patel, M.P, Hu, L, Sankaran, B, Brown, C, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2018-04-06 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Synergistic effects of functionally distinct substitutions in beta-lactamase variants shed light on the evolution of bacterial drug resistance.
J. Biol. Chem., 293, 2018
|
|
7SWI
 
 | | cTnC-TnI chimera complexed with A2 | | Descriptor: | 4-(3-cyano-3-methylazetidine-1-carbonyl)-N-[(3R,4S)-7-fluoro-4-hydroxy-6-methyl-3,4-dihydro-2H-1-benzopyran-3-yl]-5-methyl-1H-pyrrole-2-sulfonamide, Troponin C, slow skeletal and cardiac muscles,Troponin I, ... | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-19 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
6CY9
 
 | | SA11 Rotavirus NSP2 with disulfide bridge | | Descriptor: | MAGNESIUM ION, Non-structural protein 2 | | Authors: | Anish, R, Hu, L, Sankaran, B, Prasad, B.V.V. | | Deposit date: | 2018-04-05 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.615 Å) | | Cite: | Phosphorylation cascade regulates the formation and maturation of rotaviral replication factories.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7SVC
 
 | | NMR structure of cTnC-TnI chimera bound to calcium and A2 | | Descriptor: | 4-(3-cyano-3-methylazetidine-1-carbonyl)-N-[(3R,4S)-7-fluoro-4-hydroxy-6-methyl-3,4-dihydro-2H-1-benzopyran-3-yl]-5-methyl-1H-pyrrole-2-sulfonamide, CALCIUM ION, Troponin C, ... | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
7SUP
 
 | | NMR structure of cTnC-TnI chimera bound to calcium and A1 | | Descriptor: | 4-(3-cyano-3-methylazetidine-1-carbonyl)-N-[(3S)-7-fluoro-6-methyl-3,4-dihydro-2H-1-benzopyran-3-yl]-5-methyl-1H-pyrrole-2-sulfonamide, CALCIUM ION, Troponin C, ... | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-17 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
6T4I
 
 | | ROR(gamma)t ligand binding domain in complex with cholesterol and allosteric ligand MRL871 | | Descriptor: | 4-{1-[2-chloro-6-(trifluoromethyl)benzoyl]-1H-indazol-3-yl}benzoic acid, CHOLESTEROL, GLYCEROL, ... | | Authors: | de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2019-10-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Cooperativity between the orthosteric and allosteric ligand binding sites of ROR gamma t.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6T4U
 
 | | ROR(gamma)t ligand binding domain in complex with 20-alpha-hydroxycholesterol and allosteric ligand MRL871 | | Descriptor: | (3alpha,8alpha)-cholest-5-ene-3,20-diol, 4-{1-[2-chloro-6-(trifluoromethyl)benzoyl]-1H-indazol-3-yl}benzoic acid, GLYCEROL, ... | | Authors: | de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2019-10-15 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cooperativity between the orthosteric and allosteric ligand binding sites of ROR gamma t.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6T7H
 
 | | Crystal structure of Thrombin in complex with macrocycle N14-PR4-A | | Descriptor: | (14S,17R)-14-(3-carbamimidamidopropyl)-3-(furan-2-ylmethyl)-5,12,15-tris(oxidanylidene)-19-thia-3,6,13,16-tetrazatricyclo[19.4.0.0^{6,10}]pentacosa-1(25),7,9,21,23-pentaene-17-carboxamide, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Angelini, A, Kumar, M.G, Heinis, C, Cendron, L. | | Deposit date: | 2019-10-22 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Macrocycle synthesis strategy based on step-wise "adding and reacting" three components enables screening of large combinatorial libraries.
Chem Sci, 11, 2020
|
|
6T7X
 
 | | Crystal structure of PCNA from P. abyssi | | Descriptor: | DNA polymerase sliding clamp | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
6DC6
 
 | | Crystal structure of human ubiquitin activating enzyme E1 (Uba1) in complex with ubiquitin | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE 2-, Ubiquitin, ... | | Authors: | Lv, Z, Yuan, L, Williams, K.M, Atkison, J.H, Olsen, S.K. | | Deposit date: | 2018-05-04 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Crystal structure of a human ubiquitin E1-ubiquitin complex reveals conserved functional elements essential for activity.
J. Biol. Chem., 293, 2018
|
|
