2I9D
 
 | |
1U7N
 
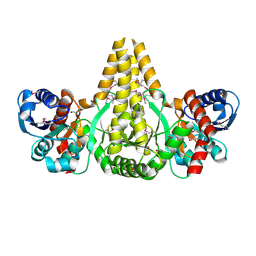 | | Crystal Structure of the fatty acid/phospholipid synthesis protein PlsX from Enterococcus faecalis V583 | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Fatty acid/phospholipid synthesis protein plsX | | Authors: | Kim, Y, Li, H, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-04 | | Release date: | 2004-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of fatty acid/phospholipid synthesis protein PlsX from Enterococcus faecalis.
J.STRUCT.FUNCT.GENOM., 10, 2009
|
|
2I7H
 
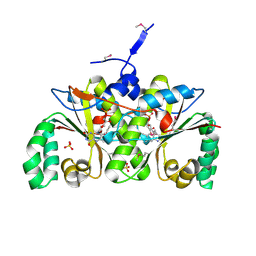 | | Crystal Structure of the Nitroreductase-like Family Protein from Bacillus cereus | | Descriptor: | FLAVIN MONONUCLEOTIDE, Nitroreductase-like family protein, SULFATE ION | | Authors: | Kim, Y, Li, H, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Nitroreductase-like Family Protein from Bacillus cereus
To be Published
|
|
2IS5
 
 | |
3SHP
 
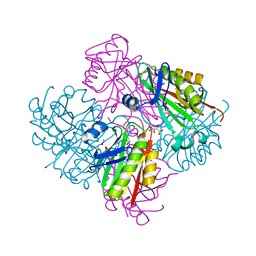 | | Crystal structure of putative acetyltransferase from Sphaerobacter thermophilus DSM 20745 | | Descriptor: | Putative acetyltransferase Sthe_0691, S,R MESO-TARTARIC ACID | | Authors: | Chang, C, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-06-16 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of putative acetyltransferase from Sphaerobacter thermophilus DSM 20745
To be Published
|
|
1TVK
 
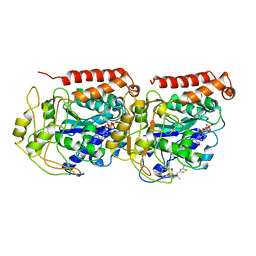 | | The binding mode of epothilone A on a,b-tubulin by electron crystallography | | Descriptor: | EPOTHILONE A, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Nettles, J.H, Li, H, Cornett, B, Krahn, J.M, Snyder, J.P, Downing, K.H. | | Deposit date: | 2004-06-29 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.89 Å) | | Cite: | The binding mode of epothilone A on alpha,beta-tubulin by electron crystallography
Science, 305, 2004
|
|
5JSG
 
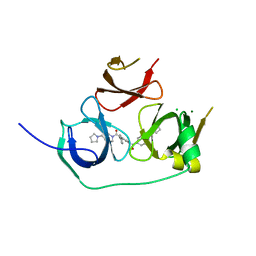 | | Crystal structure of Spindlin1 bound to compound EML405 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Spindlin-1, ... | | Authors: | Su, X, Li, H. | | Deposit date: | 2016-05-08 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Developing Spindlin1 small-molecule inhibitors by using protein microarrays
Nat. Chem. Biol., 13, 2017
|
|
5CVE
 
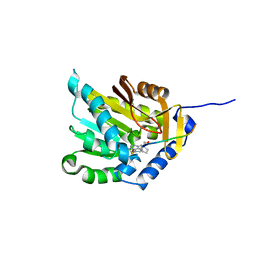 | |
9BCZ
 
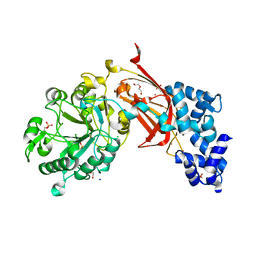 | | Chicken 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 (PLCZ1) in complex with calcium and phosphorylated threonine | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Edwards, M.M, Dong, A, Theo-Emegano, N, Seitova, A, Loppnau, P, Leung, R, Li, H, Ilyassov, O, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-10 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Chicken 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 (PLCZ1) in complex with calcium and phosphorylated threonine
To be published
|
|
5XM2
 
 | | Human N-terminal domain of FACT complex subunit SPT16 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FACT complex subunit SPT16, GLYCEROL | | Authors: | Xu, S, Li, H, Dou, Y, Chen, Y, Jiang, H, Lu, D, Wang, M, Su, D. | | Deposit date: | 2017-05-12 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | The structural basis of human Spt16 N-terminal domain interaction with histone (H3-H4)2tetramer.
Biochem.Biophys.Res.Commun., 508, 2019
|
|
5JSJ
 
 | | Crystal structure of Spindlin1 bound to compound EML631 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Spindlin-1, ... | | Authors: | Su, X, Li, H. | | Deposit date: | 2016-05-08 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Developing Spindlin1 small-molecule inhibitors by using protein microarrays
Nat. Chem. Biol., 13, 2017
|
|
9N5U
 
 | | Structure of the Thermococcus sibiricus NfnABC complex | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xiao, X, Li, H. | | Deposit date: | 2025-02-04 | | Release date: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structures define the electron bifurcating flavobicluster and ferredoxin binding site in an archaeal Nfn-Bfu transhydrogenase.
J.Biol.Chem., 301, 2025
|
|
9N5V
 
 | | Structure of the NAD(H)-bound Thermococcus sibiricus NfnABC complex | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xiao, X, Li, H. | | Deposit date: | 2025-02-04 | | Release date: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Cryo-EM structures define the electron bifurcating flavobicluster and ferredoxin binding site in an archaeal Nfn-Bfu transhydrogenase.
J.Biol.Chem., 301, 2025
|
|
4CTY
 
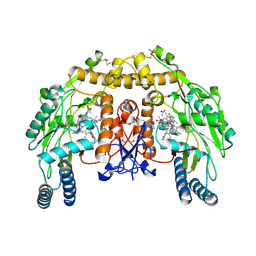 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with (R)-6-(2-Amino-2-(3-(2-(6-amino-4-methylpyridin-2-yl) ethyl)phenyl)ethyl)-4-methylpyridin-2-amine | | Descriptor: | (R)-6-(2-Amino-2-(3-(2-(6-amino-4-methylpyridin-2-yl)ethyl)phenyl)ethyl)-4-methylpyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2014-03-15 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nitric Oxide Synthase Inhibitors that Interact with Both a Heme Propionate and Tetrahydrobiopterin Show High Isoform Selectivity.
J.Med.Chem., 57, 2014
|
|
4CUM
 
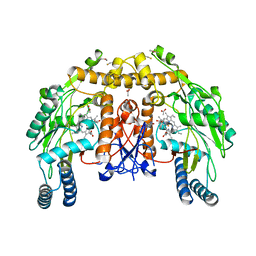 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with (9aS)-2-amino-9a-methyl-6,7,8,9,9a,10-hexahydrobenzo[g]pteridin-4(3H)-one | | Descriptor: | (9aS)-2-amino-9a-methyl-6,7,8,9,9a,10-hexahydrobenzo[g]pteridin-4(3H)-one, ACETATE ION, ARGININE, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2014-03-20 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Communication between the Zinc and Tetrahydrobiopterin Binding Sites in Nitric Oxide Synthase.
Biochemistry, 53, 2014
|
|
4CVG
 
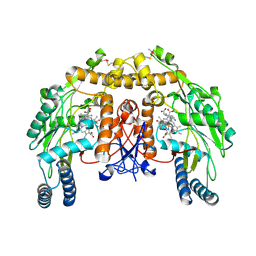 | | Structure of bovine endothelial nitric oxide synthase heme domain (H4B-free) supplemented with 50uM Zn acetate and with poor binding of 6-acetyl-2-amino-7,7-dimethyl-7,8-dihydropteridin-4(3H)-one. | | Descriptor: | ACETATE ION, ARGININE, GLYCEROL, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2014-03-25 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Communication between the Zinc and Tetrahydrobiopterin Binding Sites in Nitric Oxide Synthase.
Biochemistry, 53, 2014
|
|
4CUN
 
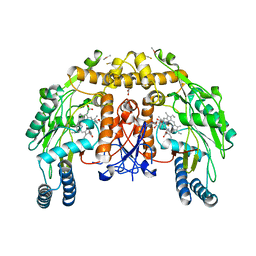 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with (9aS)-2-amino-9a-methyl-8,9,9a,10-tetrahydrobenzo[g]pteridine-4,6(3H,7H)-dione | | Descriptor: | (9aS)-2-amino-9a-methyl-8,9,9a,10-tetrahydrobenzo[g]pteridine-4,6(3H,7H)-dione, ACETATE ION, ARGININE, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2014-03-20 | | Release date: | 2014-05-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Communication between the Zinc and Tetrahydrobiopterin Binding Sites in Nitric Oxide Synthase.
Biochemistry, 53, 2014
|
|
4CTZ
 
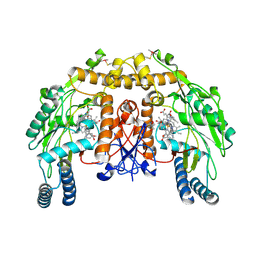 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with (S)-6-(2-amino-2-(3-(2-(4-methylpyridin-2-yl)ethyl)phenyl)ethyl)-4-methylpyridin-2-amine | | Descriptor: | (S)-6-(2-amino-2-(3-(2-(4-methylpyridin-2-yl)ethyl)phenyl)ethyl)-4-methylpyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2014-03-15 | | Release date: | 2014-05-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Nitric Oxide Synthase Inhibitors that Interact with Both a Heme Propionate and Tetrahydrobiopterin Show High Isoform Selectivity.
J.Med.Chem., 57, 2014
|
|
4CU0
 
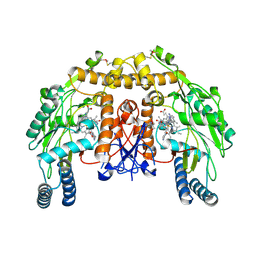 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with (R)-6-(3-amino-2-(5-(2-(6-amino-4-methylpyridin-2-yl) ethyl)pyridin-3-yl)propyl)-4-methylpyridin-2-amine | | Descriptor: | (R)-6-(3-amino-2-(5-(2-(6-amino-4-methylpyridin-2-yl)ethyl)pyridin-3-yl)propyl)-4-methylpyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2014-03-15 | | Release date: | 2014-05-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Nitric Oxide Synthase Inhibitors that Interact with Both a Heme Propionate and Tetrahydrobiopterin Show High Isoform Selectivity.
J.Med.Chem., 57, 2014
|
|
4D3A
 
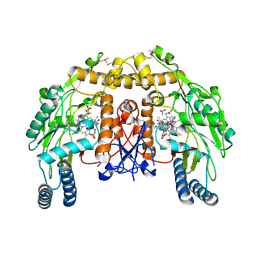 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 3-(3-fluorophenyl)-N-2-(2-(5-methyl-1H-imidazol-1-yl) pyrimidin-4-yl)ethylpropan-1-amine | | Descriptor: | 3-(3-fluorophenyl)-N-{2-[2-(5-methyl-1H-imidazol-1-yl)pyrimidin-4-yl]ethyl}propan-1-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2014-10-20 | | Release date: | 2014-12-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Novel 2,4-Disubstituted Pyrimidines as Potent, Selective, and Cell-Permeable Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
4W5A
 
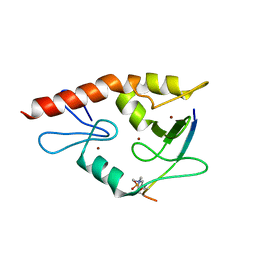 | | Complex structure of ATRX ADD bound to H3K9me3S10ph peptide | | Descriptor: | Peptide from Histone H3.3, Transcriptional regulator ATRX, ZINC ION | | Authors: | Zhao, D, Xiang, B, Li, H. | | Deposit date: | 2014-08-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | ATRX tolerates activity-dependent histone H3 methyl/phos switching to maintain repetitive element silencing in neurons
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5XUR
 
 | | Crystal Structure of Rv2466c C22S Mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Thioredoxin-like reductase Rv2466c | | Authors: | Zhang, X, Li, H. | | Deposit date: | 2017-06-25 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Identification of a Mycothiol-Dependent Nitroreductase from Mycobacterium tuberculosis.
ACS Infect Dis, 4, 2018
|
|
9NEA
 
 | | Human polymerase epsilon bound to PCNA and DNA with a pre-existing mismatch in the blocked conformation II | | Descriptor: | DNA (29-MER), DNA (35-MER), DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2025-02-19 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | The proofreading mechanism of the human leading-strand DNA polymerase epsilon holoenzyme.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9NE9
 
 | | Human polymerase epsilon bound to PCNA and DNA with a pre-existing mismatch in the blocked conformation I | | Descriptor: | DNA (35-MER), DNA (49-MER), DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2025-02-19 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | The proofreading mechanism of the human leading-strand DNA polymerase epsilon holoenzyme.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9NE7
 
 | | Human polymerase epsilon bound to PCNA and DNA with an in-situ-generated mismatch in the Pol-backtracking state | | Descriptor: | DNA (33-MER), DNA (47-MER), DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2025-02-19 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | The proofreading mechanism of the human leading-strand DNA polymerase epsilon holoenzyme.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
