8FLM
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP, NVS-STG2 and C53 | | Descriptor: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, ... | | Authors: | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
2GIC
 
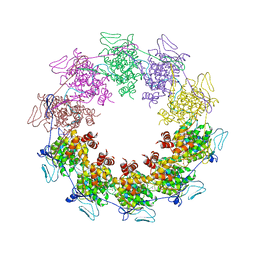 | | Crystal Structure of a vesicular stomatitis virus nucleocapsid-RNA complex | | Descriptor: | 45-MER, Nucleocapsid protein, URANYL (VI) ION | | Authors: | Green, T.J, Zhang, X, Wertz, G.W, Luo, M. | | Deposit date: | 2006-03-28 | | Release date: | 2006-08-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structure of the vesicular stomatitis virus nucleoprotein-RNA complex unveils how the RNA is sequestered
Science, 313, 2006
|
|
1WOV
 
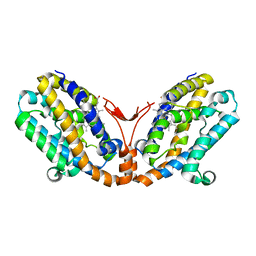 | | Crystal structure of heme oxygenase-2 from Synechocystis sp. PCC 6803 in complex with heme | | Descriptor: | Heme oxygenase 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Hagiwara, Y, Zhang, X, Yoshida, T, Migita, C.T, Fukuyama, K. | | Deposit date: | 2004-08-26 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of dimeric heme oxygenase-2 from Synechocystis sp. PCC 6803 in complex with heme.
Biochemistry, 44, 2005
|
|
1WOW
 
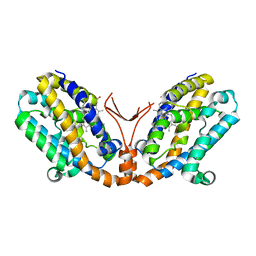 | | Crystal structure of heme oxygenase-2 from Synechocystis sp. PCC 6803 complexed with heme in ferrous form | | Descriptor: | Heme oxygenase 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Hagiwara, Y, Zhang, X, Yoshida, T, Migita, C.T, Fukuyama, K. | | Deposit date: | 2004-08-26 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of dimeric heme oxygenase-2 from Synechocystis sp. PCC 6803 in complex with heme.
Biochemistry, 44, 2005
|
|
3PUS
 
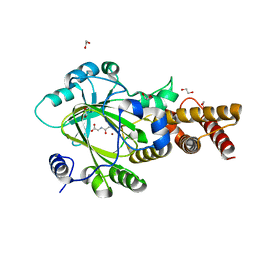 | | PHF2 Jumonji-NOG-Ni(II) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-OXALYLGLYCINE, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2010-12-06 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis for human PHF2 Jumonji domain interaction with metal ions.
J.Mol.Biol., 406, 2011
|
|
3PU8
 
 | | PHF2 Jumonji-NOG-Fe(II) complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2010-12-03 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Structural basis for human PHF2 Jumonji domain interaction with metal ions.
J.Mol.Biol., 406, 2011
|
|
3PTR
 
 | | PHF2 Jumonji domain | | Descriptor: | 1,2-ETHANEDIOL, PHD finger protein 2 | | Authors: | Horton, J.R, Upadhyay, A.K, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2010-12-03 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structural basis for human PHF2 Jumonji domain interaction with metal ions.
J.Mol.Biol., 406, 2011
|
|
3PUA
 
 | | PHF2 Jumonji-NOG-Ni(II) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-OXALYLGLYCINE, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2010-12-03 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for human PHF2 Jumonji domain interaction with metal ions.
J.Mol.Biol., 406, 2011
|
|
3PU3
 
 | | PHF2 Jumonji domain-NOG complex | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-OXALYLGLYCINE, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2010-12-03 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for human PHF2 Jumonji domain interaction with metal ions.
J.Mol.Biol., 406, 2011
|
|
1WOX
 
 | | Crystal structure of heme oxygenase-2 from Synechocystis sp. PCC 6803 in complex with heme and NO | | Descriptor: | Heme oxygenase 2, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Hagiwara, Y, Zhang, X, Yoshida, T, Migita, C.T, Fukuyama, K. | | Deposit date: | 2004-08-26 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of dimeric heme oxygenase-2 from Synechocystis sp. PCC 6803 in complex with heme.
Biochemistry, 44, 2005
|
|
2J48
 
 | |
8WJL
 
 | | Cryo-EM structure of 6-subunit Smc5/6 hinge region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.15 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8WJO
 
 | | Cryo-EM structure of 8-subunit Smc5/6 arm region | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.04 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8WJN
 
 | | Cryo-EM structure of 6-subunit Smc5/6 head region | | Descriptor: | Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosomes element 1, Non-structural maintenance of chromosomes element 4, ... | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.58 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
2IPA
 
 | | solution structure of Trx-ArsC complex | | Descriptor: | Protein arsC, Thioredoxin | | Authors: | Jin, C, Hu, Y, Li, Y, Zhang, X. | | Deposit date: | 2006-10-12 | | Release date: | 2007-02-13 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Conformational fluctuations coupled to the thiol-disulfide transfer between thioredoxin and arsenate reductase in Bacillus subtilis.
J.Biol.Chem., 282, 2007
|
|
8H3G
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166V Mutant in Complex with Inhibitor Enstrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, GLYCEROL | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
7WB4
 
 | | Cryo-EM structure of the NR subunit from X. laevis NPC | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Huang, G, Zhan, X, Shi, Y. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Cryo-EM structure of the nuclear ring from Xenopus laevis nuclear pore complex.
Cell Res., 32, 2022
|
|
8H3L
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (T21I and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3K
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, ... | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
3UT4
 
 | | Structural view of a non Pfam singleton and crystal packing analysis | | Descriptor: | Putative uncharacterized protein | | Authors: | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
3UT7
 
 | | Structural view of a non Pfam singleton and crystal packing analysis | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
8IAB
 
 | | The Arabidopsis CLCa transporter bound with chloride, ATP and PIP2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Chloride channel protein CLC-a, ... | | Authors: | Yang, Z, Zhang, X, Zhang, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Molecular mechanism underlying regulation of Arabidopsis CLCa transporter by nucleotides and phospholipids.
Nat Commun, 14, 2023
|
|
8IAD
 
 | | The Arabidopsis CLCa transporter bound with nitrate, ATP and PIP2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chloride channel protein CLC-a, MAGNESIUM ION, ... | | Authors: | Yang, Z, Zhang, X, Zhang, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Molecular mechanism underlying regulation of Arabidopsis CLCa transporter by nucleotides and phospholipids.
Nat Commun, 14, 2023
|
|
8Y8C
 
 | | Structure of HCoV-HKU1C spike in the inactive-closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8D
 
 | | Structure of HCoV-HKU1C spike in the inactive-1up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
