3M79
 
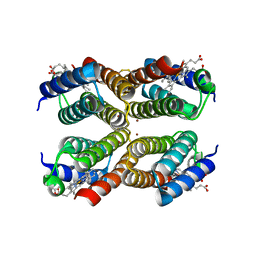 | |
4ARR
 
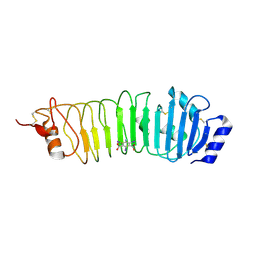 | | Crystal structure of the N-terminal domain of Drosophila Toll receptor with the magic triangle I3C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, TOLL RECEPTOR, ... | | Authors: | Gangloff, M, Gay, N.J. | | Deposit date: | 2012-04-26 | | Release date: | 2013-02-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Liesegang-Like Patterns of Toll Crystals Grown in Gel.
J.Appl.Crystallogr., 46, 2013
|
|
5Y95
 
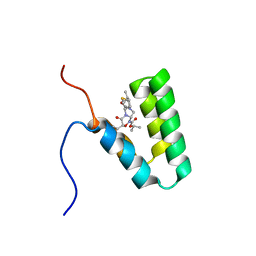 | | Haddock model of mSIN3B PAH1 domain | | Descriptor: | Paired amphipathic helix protein Sin3b, propan-2-yl (3R,6S,9aS)-3-ethyl-8-(3-methylbutyl)-6-(2-methylsulfanylethyl)-4,7-bis(oxidanylidene)-9,9a-dihydro-6H-pyrazino[2,1-c][1,2,4]oxadiazine-1-carboxylate | | Authors: | Kurita, J, Hirao, Y, Nishimura, Y. | | Deposit date: | 2017-08-22 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A mimetic of the mSin3-binding helix of NRSF/REST ameliorates abnormal pain behavior in chronic pain models.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
3D30
 
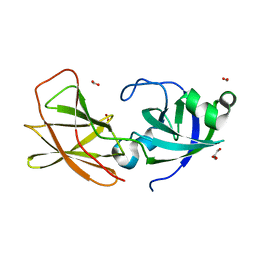 | | Structure of an expansin like protein from Bacillus Subtilis at 1.9A resolution | | Descriptor: | Expansin like protein, FORMIC ACID, GLYCEROL | | Authors: | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Joris, B, Charlier, P. | | Deposit date: | 2008-05-09 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and activity of Bacillus subtilis YoaJ (EXLX1), a bacterial expansin that promotes root colonization.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2MN2
 
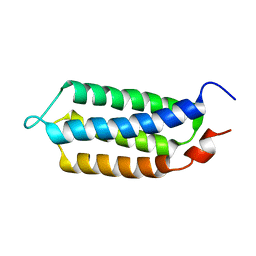 | | 3D structure of YmoB, a modulator of biofilm formation | | Descriptor: | YmoB | | Authors: | Marimon, O, Cordeiro, T.N, Amata, I, Pons, M. | | Deposit date: | 2014-03-26 | | Release date: | 2015-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An oxygen-sensitive toxin-antitoxin system.
Nat Commun, 7, 2016
|
|
5O59
 
 | | Cellobiohydrolase Cel7A from T. atroviride | | Descriptor: | 1-thio-beta-D-glucopyranose, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Borisova, A.S, Stahlberg, J, Hansson, H. | | Deposit date: | 2017-06-01 | | Release date: | 2018-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Correlation of structure, function and protein dynamics in GH7 cellobiohydrolases from Trichoderma atroviride, T. reesei and T. harzianum.
Biotechnol Biofuels, 11, 2018
|
|
5O5D
 
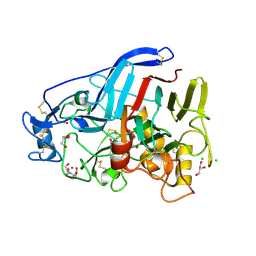 | | Cellobiohydrolase Cel7A from T. atroviride | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Borisova, A.S, Stahlberg, J, Hansson, H. | | Deposit date: | 2017-06-01 | | Release date: | 2018-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Correlation of structure, function and protein dynamics in GH7 cellobiohydrolases from Trichoderma atroviride, T. reesei and T. harzianum.
Biotechnol Biofuels, 11, 2018
|
|
1Q38
 
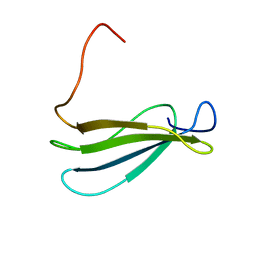 | | Anastellin | | Descriptor: | Fibronectin | | Authors: | Briknarova, K, Akerman, M.E, Hoyt, D.W, Ruoslahti, E, Ely, K.R. | | Deposit date: | 2003-07-28 | | Release date: | 2003-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Anastellin, an FN3 fragment with fibronectin polymerization activity, resembles amyloid fibril precursors
J.Mol.Biol., 332, 2003
|
|
4ARN
 
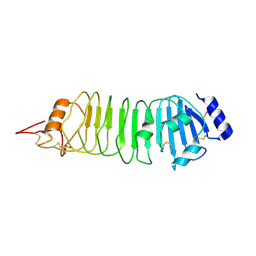 | | Crystal structure of the N-terminal domain of Drosophila Toll receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MALONATE ION, ... | | Authors: | Gangloff, M, Gay, N.J. | | Deposit date: | 2012-04-25 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Functional Insights from the Crystal Structure of the N-Terminal Domain of the Prototypical Toll Receptor.
Structure, 21, 2013
|
|
2ZN7
 
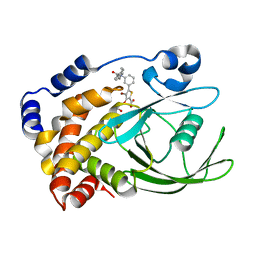 | | CRYSTAL STRUCTURES OF PTP1B-Inhibitor Complexes | | Descriptor: | 4-bromo-3-(carboxymethoxy)-5-{3-[cyclohexyl(phenylcarbonyl)amino]phenyl}thiophene-2-carboxylic acid, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W, Wu, J. | | Deposit date: | 2008-04-22 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based optimization of protein tyrosine phosphatase-1 B inhibitors: capturing interactions with arginine 24
Chemmedchem, 3, 2008
|
|
3AII
 
 | |
3SJT
 
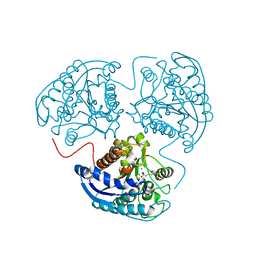 | | Crystal structure of human arginase I in complex with the inhibitor Me-ABH, Resolution 1.60 A, twinned structure | | Descriptor: | Arginase-1, MANGANESE (II) ION, [(5S)-5-amino-5-carboxyhexyl](trihydroxy)borate | | Authors: | Di Costanzo, L, Christianson, D.W. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Binding of alpha , alpha-disubstituted amino acids to arginase suggests new avenues for inhibitor design.
J.Med.Chem., 54, 2011
|
|
2VWU
 
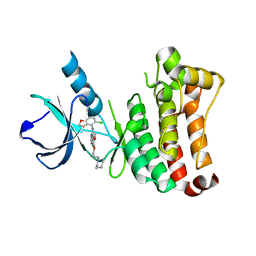 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, N-(5-chloro-1,3-benzodioxol-4-yl)-6-methoxy-7-(3-piperidin-1-ylpropoxy)quinazolin-4-amine | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Rowsell, S, Packer, M, McAlister, M. | | Deposit date: | 2008-06-27 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 1: Structure-Based Design and Optimization of a Series of 2,4-Bis-Anilinopyrimidines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VWV
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, N'-(3-CHLORO-4-METHOXY-PHENYL)-N-(3,4,5-TRIMETHOXYPHENYL)-1,3,5-TRIAZINE-2,4-DIAMINE | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Kettle, J.G, Leach, A.G. | | Deposit date: | 2008-06-27 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 1: Structure-Based Design and Optimization of a Series of 2,4-Bis-Anilinopyrimidines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2ZMM
 
 | | Crystal structure of PTP1B-inhibitor complex | | Descriptor: | 4-bromo-3-(carboxymethoxy)-5-{3-[cyclohexyl(methylcarbamoyl)amino]phenyl}thiophene-2-carboxylic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Xu, W, Wu, J. | | Deposit date: | 2008-04-19 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based optimization of protein tyrosine phosphatase-1 B inhibitors: capturing interactions with arginine 24
Chemmedchem, 3, 2008
|
|
1RT8
 
 | | CRYSTAL STRUCTURE OF THE ACTIN-CROSSLINKING CORE OF SCHIZOSACCHAROMYCES POMBE FIMBRIN | | Descriptor: | SULFATE ION, fimbrin | | Authors: | Klein, M.G, Shi, W, Ramagopal, U, Tseng, Y, Wirtz, D, Kovar, D.R, Staiger, C.J, Almo, S.C. | | Deposit date: | 2003-12-10 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the actin crosslinking core of fimbrin.
Structure, 12, 2004
|
|
2VWW
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, N'-(5-CHLORO-1,3-BENZODIOXOL-4-YL)-N-(3,4,5- TRIMETHOXYPHENYL)PYRIMIDINE-2,4-DIAMINE | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | Deposit date: | 2008-06-27 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 1: Structure-Based Design and Optimization of a Series of 2,4-Bis-Anilinopyrimidines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
