7SJR
 
 | | Cryo-EM structure of AdnA-AdnB(W325A) in complex with DNA and AMPPNP | | Descriptor: | DNA (70-MER), DNA helicase, IRON/SULFUR CLUSTER, ... | | Authors: | Wang, J, Warren, G.M, Shuman, S, Patel, D.J. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-activity relationships at a nucleobase-stacking tryptophan required for chemomechanical coupling in the DNA resecting motor-nuclease AdnAB.
Nucleic Acids Res., 50, 2022
|
|
3TTI
 
 | | Crystal Structure of JNK3 complexed with CC-930, an orally active anti-fibrotic JNK inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 10, trans-4-({9-[(3S)-tetrahydrofuran-3-yl]-8-[(2,4,6-trifluorophenyl)amino]-9H-purin-2-yl}amino)cyclohexanol | | Authors: | Plantevin-Krenitsky, V, Nadolny, L, Delgado, M, Ayala, L, Clareen, S, Hilgraf, R, Albers, R, Hegde, S, D'Sidocky, N, Sapienza, J, Wright, J, McCarrick, M, Bahmanyar, S, Chamberlain, P, Delker, S.L, Muir, J, Giegel, D, Xu, L, Celeridad, M, Lachowitzer, J, Bennett, B, Moghaddam, M, Khatsenko, O, Katz, J, Fan, R, Bai, A, Tang, Y, Shirley, M.A, Benish, B, Bodine, T, Blease, K, Raymon, H, Cathers, B.E, Satoh, Y. | | Deposit date: | 2011-09-14 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of CC-930, an orally active anti-fibrotic JNK inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5DZX
 
 | | Protocadherin beta 6 extracellular cadherin domains 1-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin beta 6, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2015-09-26 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.879 Å) | | Cite: | Structural Basis of Diverse Homophilic Recognition by Clustered alpha- and beta-Protocadherins.
Neuron, 90, 2016
|
|
8OOK
 
 | |
8OO9
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-DNA refinement state1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase INO80, DNA strand 1, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOA
 
 | | CryoEM Structure INO80core Hexasome complex Hexasome refinement state1 | | Descriptor: | DNA Strand 2, DNA strand 1, Histone H2A, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOF
 
 | | CryoEM Structure INO80core Hexasome complex Arp5 Ies6 refinement state1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling complex subunit IES6, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
1MVF
 
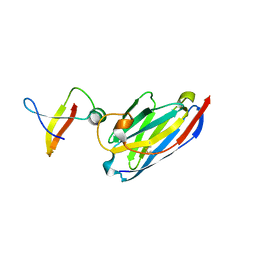 | | MazE addiction antidote | | Descriptor: | PemI-like protein 1, immunoglobulin heavy chain variable region | | Authors: | Loris, R, Marianovsky, I, Lah, J, Laeremans, T, Engelberg-Kulka, H, Glaser, G, Muyldermans, S, Wyns, L. | | Deposit date: | 2002-09-25 | | Release date: | 2003-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the intrinsically flexible addiction antidote MazE.
J.Biol.Chem., 278, 2003
|
|
5BVN
 
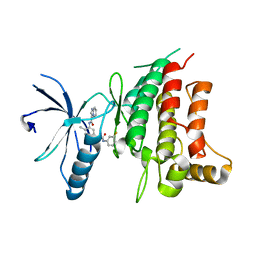 | | Fragment-based discovery of potent and selective DDR1/2 inhibitors | | Descriptor: | Epithelial discoidin domain-containing receptor 1, IODIDE ION, N-[5-({[(3-fluorophenyl)carbamoyl]amino}methyl)-2-methylphenyl]imidazo[1,2-a]pyridine-3-carboxamide | | Authors: | Murray, C, Berdini, V, Buck, I, Carr, M, Cleasby, A, Coyle, J, Curry, J, Day, J, Hearn, K, Iqbal, A, Lee, L, Martins, V, Mortenson, P, Munck, J, Page, L, Patel, S, Roomans, S, Kirsten, T, Saxty, G. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Fragment-Based Discovery of Potent and Selective DDR1/2 Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
2OBO
 
 | | Structure of HEPATITIS C VIRAL NS3 protease domain complexed with NS4A peptide and ketoamide SCH476776 | | Descriptor: | BETA-MERCAPTOETHANOL, HCV NS3 protease, HCV NS4A peptide, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
1NBO
 
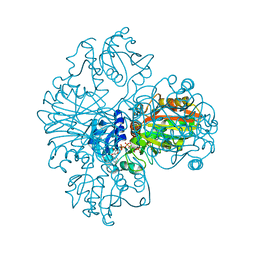 | | The dual coenzyme specificity of photosynthetic glyceraldehyde-3-phosphate dehydrogenase interpreted by the crystal structure of A4 isoform complexed with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, glyceraldehyde-3-phosphate dehydrogenase A | | Authors: | Falini, G, Fermani, S, Ripamonti, A, Sabatino, P, Sparla, F, Pupillo, P, Trost, P. | | Deposit date: | 2002-12-03 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dual Coenzyme Specificity of Photosynthetic Glyceraldehyde-3-phosphate
Dehydrogenase Interpreted by the Crystal Structure of A(4) Isoform
Complexed with NAD
Biochemistry, 42, 2003
|
|
3I0T
 
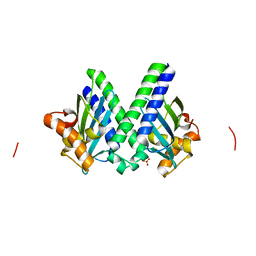 | | Sulfur-SAD at long wavelength: Structure of BH3703 from Bacillus halodurans | | Descriptor: | BH3703 protein, SULFATE ION | | Authors: | Ramagopal, U.A, Toro, R, Wasserman, S, Burley, S.K, Almo, S.C. | | Deposit date: | 2009-06-25 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Sulfur-SAD at long wavelength: Structure of BH3703 from Bacillus halodurans
To be published
|
|
3E61
 
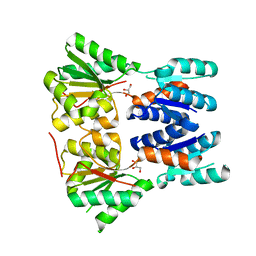 | | Crystal structure of a putative transcriptional repressor of ribose operon from Staphylococcus saprophyticus subsp. saprophyticus | | Descriptor: | GLYCEROL, Putative transcriptional repressor of ribose operon | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-14 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative transcriptional repressor of ribose operon from Staphylococcus saprophyticus subsp. saprophyticus
To be Published
|
|
6O0X
 
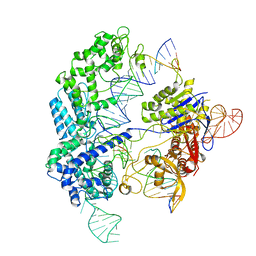 | | Conformational states of Cas9-sgRNA-DNA ternary complex in the presence of magnesium | | Descriptor: | 3' product of target strand DNA, 5' product of target strand DNA, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Zhu, X, Clarke, R, Puppala, A.K, Chittori, S, Merk, A, Merrill, B.J, Simonovic, M, Subramaniam, S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2MUS
 
 | | HADDOCK calculated model of LIN5001 bound to the HET-s amyloid | | Descriptor: | 3''',4'-bis(carboxymethyl)-2,2':5',2'':5'',2''':5''',2''''-quinquethiophene-5,5''''-dicarboxylic acid, Heterokaryon incompatibility protein s | | Authors: | Hermann, U.S, Schuetz, A.K, Shirani, H, Saban, D, Nuvolone, M, Huang, D.H, Li, B, Ballmer, B, Aslund, A.K.O, Mason, J.J, Rushing, E, Budka, H, Hammarstrom, P, Bockmann, A, Caflisch, A, Meier, B.H, Nilsson, P.K.R, Hornemann, S, Aguzzi, A. | | Deposit date: | 2014-09-16 | | Release date: | 2017-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-based drug design identifies polythiophenes as antiprion compounds.
Sci Transl Med, 7, 2015
|
|
3E3M
 
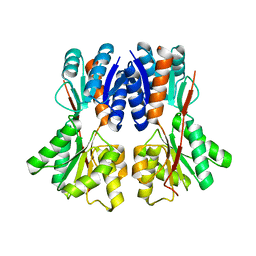 | | Crystal structure of a LacI family transcriptional regulator from Silicibacter pomeroyi | | Descriptor: | Transcriptional regulator, LacI family | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Iizuka, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-07 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a LacI family transcriptional regulator from Silicibacter pomeroyi
To be Published
|
|
3ER6
 
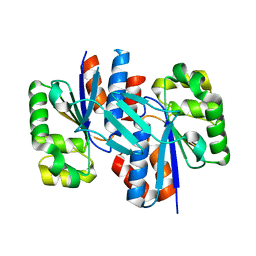 | | Crystal structure of a putative transcriptional regulator protein from Vibrio parahaemolyticus | | Descriptor: | Putative transcriptional regulator protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative transcriptional regulator protein from Vibrio parahaemolyticus
To be Published
|
|
3IJM
 
 | | The structure of a restriction endonuclease-like fold superfamily protein from Spirosoma linguale. | | Descriptor: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Cuff, M.E, Tesar, C, Samano, S, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-04 | | Release date: | 2009-09-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of a restriction endonuclease-like fold superfamily protein from Spirosoma linguale.
TO BE PUBLISHED
|
|
3FK9
 
 | | Crystal structure of mMutator MutT protein from Bacillus halodurans | | Descriptor: | Mutator MutT protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Hu, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-16 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of mMutator MutT protein from Bacillus halodurans
To be Published
|
|
3IF6
 
 | | Crystal structure of OXA-46 beta-lactamase from P. aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, HEXAETHYLENE GLYCOL, L(+)-TARTARIC ACID, ... | | Authors: | Docquier, J.D, Benvenuti, M, Calderone, V, Giuliani, F, Kapetis, D, De Luca, F, Rossolini, G.M, Mangani, S. | | Deposit date: | 2009-07-24 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the narrow-spectrum OXA-46 class D beta-lactamase: relationship between active-site lysine carbamylation and inhibition by polycarboxylates
Antimicrob.Agents Chemother., 54, 2010
|
|
6X5P
 
 | | Discovery of Hydroxy Pyrimidine Factor IXa Inhibitors | | Descriptor: | 3-chloro-4-{[5-hydroxy-6-(4-methylphenyl)pyrimidin-4-yl]amino}benzene-1-carboximidamide, CITRIC ACID, Coagulation factor IX, ... | | Authors: | Jayne, C.L, Andreani, T, Chan, T, Chelliah, M.V, Clasby, M.C, Dwyer, M, Eagen, K.A, Fried, S, Greenlee, W.J, Guo, Z, Hawes, B, Hruza, A, Ingram, R, Keertikar, K.M, Neelamkavil, S, Reichert, P, Xia, Y, Chackalamannil, S. | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Discovery of hydroxy pyrimidine Factor IXa inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6O0Y
 
 | | Conformational states of Cas9-sgRNA-DNA ternary complex in the presence of magnesium | | Descriptor: | 3' product of target strand DNA, 5' product of target strand DNA, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Zhu, X, Clarke, R, Puppala, A.K, Chittori, S, Merk, A, Merrill, B.J, Simonovic, M, Subramaniam, S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3EYP
 
 | | Crystal structure of putative alpha-L-fucosidase from Bacteroides thetaiotaomicron | | Descriptor: | GLYCEROL, Putative alpha-L-fucosidase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Hu, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-21 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative alpha-L-fucosidase from Bacteroides thetaiotaomicron
To be Published
|
|
8AGI
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) in complex with JMC31 | | Descriptor: | 1-[2,4-bis(oxidanyl)-5-propan-2-yl-phenyl]-5-[4-[(cyclohexylmethylamino)methyl]phenyl]-~{N}-ethyl-1,2,3-triazole-4-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Tassone, G, Pozzi, C, Mazzorana, M, Mangani, S, Maramai, S. | | Deposit date: | 2022-07-20 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of Human Heat Shock Protein 90 N-Terminal Domain and Its Variants K112R and K112A in Complex with a Potent 1,2,3-Triazole-Based Inhibitor.
Int J Mol Sci, 23, 2022
|
|
8AGJ
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) variant K112A in complex with JMC31 | | Descriptor: | 1-[2,4-bis(oxidanyl)-5-propan-2-yl-phenyl]-5-[4-[(cyclohexylmethylamino)methyl]phenyl]-~{N}-ethyl-1,2,3-triazole-4-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Tassone, G, Pozzi, C, Mazzorana, M, Mangani, S, Maramai, S. | | Deposit date: | 2022-07-20 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Characterization of Human Heat Shock Protein 90 N-Terminal Domain and Its Variants K112R and K112A in Complex with a Potent 1,2,3-Triazole-Based Inhibitor.
Int J Mol Sci, 23, 2022
|
|
