7DUU
 
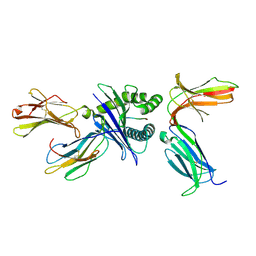 | | Crystal structure of HLA molecule with an KIR receptor | | Descriptor: | Beta-2-microglobulin, Killer cell immunoglobulin-like receptor 2DS2, LEU-ASN-PRO-SER-VAL-ALA-ALA-THR-LEU, ... | | Authors: | Yang, Y, Yin, L. | | Deposit date: | 2021-01-11 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Activating receptor KIR2DS2 bound to HLA-C1 reveals the novel recognition features of activating receptor.
Immunology, 165, 2022
|
|
7WHD
 
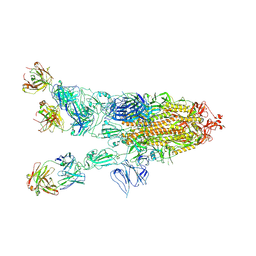 | | SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (2u1d) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zeng, J.W, Wang, X.W, Ge, J.W, Wang, Z.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | SARS-CoV-2 hijacks neutralizing dimeric IgA for nasal infection and injury in Syrian hamsters 1 .
Emerg Microbes Infect, 12, 2023
|
|
4ERE
 
 | | crystal structure of MDM2 (17-111) in complex with compound 23 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(3R,5R,6S)-1-[(2S)-1-tert-butoxy-1-oxobutan-2-yl]-5-(3-chlorophenyl)-6-(4-chlorophenyl)-2-oxopiperidin-3-yl]acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
7WH8
 
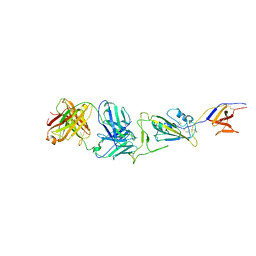 | |
7WHB
 
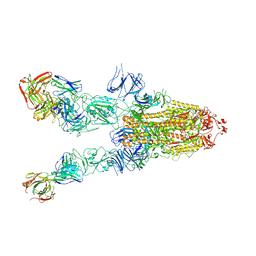 | | SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (3U) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zeng, J.W, Ge, J.W, Wang, X.Q. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | SARS-CoV-2 hijacks neutralizing dimeric IgA for nasal infection and injury in Syrian hamsters 1 .
Emerg Microbes Infect, 12, 2023
|
|
5X5M
 
 | |
5HCV
 
 | |
4ERF
 
 | | crystal structure of MDM2 (17-111) in complex with compound 29 (AM-8553) | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, {(3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-1-[(2S,3S)-2-hydroxypentan-3-yl]-3-methyl-2-oxopiperidin-3-yl}acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
8F8Y
 
 | | PHF2 (PHD+JMJ) in Complex with VRK1 N-Terminal Peptide | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase PHF2, SULFATE ION, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2022-11-22 | | Release date: | 2023-01-18 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | A complete methyl-lysine binding aromatic cage constructed by two domains of PHF2.
J.Biol.Chem., 299, 2022
|
|
8F8Z
 
 | |
5XFS
 
 | |
4EQK
 
 | |
4ENF
 
 | | Crystal structure of the cap-binding domain of polymerase basic protein 2 from influenza virus A/Puerto Rico/8/34(h1n1) | | Descriptor: | 1,4-BUTANEDIOL, NITRATE ION, Polymerase basic protein 2 | | Authors: | Meng, G, Liu, Y, Zheng, X. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural and functional characterization of K339T substitution identified in the PB2 subunit cap-binding pocket of influenza A virus
J.Biol.Chem., 288, 2013
|
|
4HAC
 
 | |
7E9Q
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(1 out RBD, state3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7E9O
 
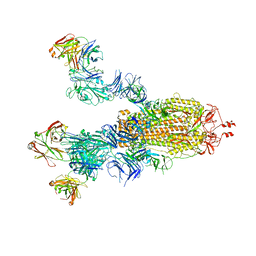 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(3 up RBDs, state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7E9P
 
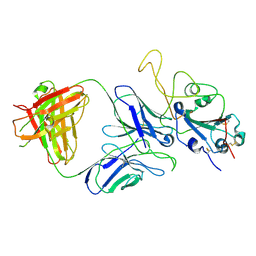 | |
7E9N
 
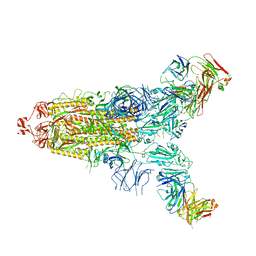 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(1 down RBD, state1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7ENF
 
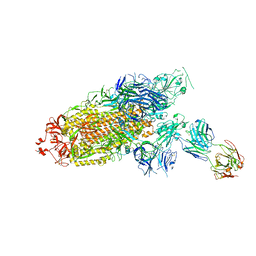 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with Fab30 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab30, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7ENG
 
 | |
8H3T
 
 | | The crystal structure of AlpH | | Descriptor: | AlpH, GLYCEROL | | Authors: | Zhao, Y, Li, M, Jiang, M, Pan, L.F. | | Deposit date: | 2022-10-09 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.866 Å) | | Cite: | O-methyltransferase-like enzyme catalyzed diazo installation in polyketide biosynthesis.
Nat Commun, 14, 2023
|
|
8OYS
 
 | | De novo designed TIM barrel fold TBF_24 | | Descriptor: | CHLORIDE ION, De novo designed TIM-barrel | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Computational design of soluble and functional membrane protein analogues.
Nature, 631, 2024
|
|
8OYV
 
 | | De novo designed Claudin fold CLF_4 | | Descriptor: | De novo designed soluble Claudin | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Computational design of soluble and functional membrane protein analogues.
Nature, 631, 2024
|
|
8OYX
 
 | | De novo designed soluble GPCR-like fold GLF_18 | | Descriptor: | De novo designed soluble GPCR-like protein, PHOSPHATE ION | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Computational design of soluble and functional membrane protein analogues.
Nature, 631, 2024
|
|
8OYY
 
 | | De novo designed soluble GPCR-like fold GLF_32 | | Descriptor: | CHLORIDE ION, De novo designed soluble GPCR-like protein, POTASSIUM ION | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Computational design of soluble and functional membrane protein analogues.
Nature, 631, 2024
|
|
