8SLY
 
 | | Rat TRPV2 bound with 2 CBD ligands in nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, Transient receptor potential cation channel subfamily V member 2, ... | | Authors: | Tan, X, Swartz, K.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cannabidiol sensitizes TRPV2 channels to activation by 2-APB.
Elife, 12, 2023
|
|
7DK0
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW05 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MW05 heavy chain, MW05 light chain, ... | | Authors: | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-11-22 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires Fc gamma RIIB and virus-antibody complex with bivalent interaction.
Commun Biol, 5, 2022
|
|
7DJZ
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW01 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, MW01 heavy chain, ... | | Authors: | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-11-22 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires Fc gamma RIIB and virus-antibody complex with bivalent interaction.
Commun Biol, 5, 2022
|
|
7V6F
 
 | | Structure of Candida albicans Fructose-1,6-bisphosphate aldolase complexed with G3P | | Descriptor: | Fructose-bisphosphate aldolase, GLYCERALDEHYDE-3-PHOSPHATE, ZINC ION | | Authors: | Hongxuan, C, Huang, Y, Han, C, Chen, W, Ren, Y, Wan, J. | | Deposit date: | 2021-08-20 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
7XML
 
 | | Cryo-EM structure of PEIP-Bs_enolase complex | | Descriptor: | Enolase, MAGNESIUM ION, Putative gene 60 protein | | Authors: | Li, S, Zhang, K. | | Deposit date: | 2022-04-26 | | Release date: | 2022-07-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Bacteriophage protein PEIP is a potent Bacillus subtilis enolase inhibitor.
Cell Rep, 40, 2022
|
|
3M37
 
 | |
7LZ7
 
 | | Tubulin-RB3_SLD-TTL in complex with compound 5k | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(3,6-dimethyl[1,2]oxazolo[5,4-d]pyrimidin-4-yl)-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
7LZ8
 
 | | Tubulin-RB3_SLD-TTL in complex with compound 5t | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[2-(ethylamino)pyrido[3,2-d]pyrimidin-4-yl]-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
3MOO
 
 | | Crystal structure of the HmuO, heme oxygenase from Corynebacterium diphtheriae, in complex with azide-bound verdoheme | | Descriptor: | 5-OXA-PROTOPORPHYRIN IX CONTAINING FE, AZIDE ION, Heme oxygenase, ... | | Authors: | Omori, K, Matsui, T, Unno, M, Ikeda-Saito, M. | | Deposit date: | 2010-04-22 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Enzymatic ring-opening mechanism of verdoheme by the heme oxygenase: a combined X-ray crystallography and QM/MM study.
J.Am.Chem.Soc., 132, 2010
|
|
8TGN
 
 | | VMAT1 dimer with serotonin and reserpine | | Descriptor: | Chromaffin granule amine transporter, SEROTONIN, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
8TGL
 
 | | VMAT1 dimer with norepinephrine and reserpine | | Descriptor: | Chromaffin granule amine transporter, L-NOREPINEPHRINE, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
8TGI
 
 | | VMAT1 dimer with dopamine and reserpine | | Descriptor: | Chromaffin granule amine transporter, L-DOPAMINE, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
8TGM
 
 | | VMAT1 dimer with reserpine | | Descriptor: | VMAT1 dimer with reserpine, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
8TGG
 
 | | VMAT1 dimer with MPP+ and reserpine | | Descriptor: | 1-methyl-4-phenylpyridin-1-ium, Chromaffin granule amine transporter, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
8TGH
 
 | | VMAT1 dimer with amphetamine and reserpine | | Descriptor: | (2S)-1-phenylpropan-2-amine, Chromaffin granule amine transporter, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
8TGK
 
 | | VMAT1 dimer with histamine and reserpine | | Descriptor: | Chromaffin granule amine transporter, HISTAMINE, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
8TGJ
 
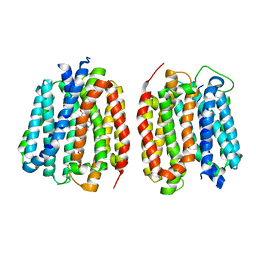 | | VMAT1 dimer in unbound form and with reserpine | | Descriptor: | VMAT1 dimer in unbound form and with reserpine, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
8TZS
 
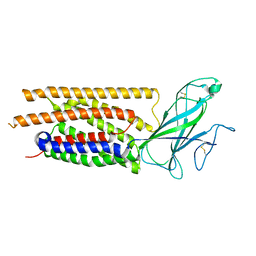 | | Structure of human WLS | | Descriptor: | Protein wntless homolog | | Authors: | Qi, X, Hu, Q, Li, X. | | Deposit date: | 2023-08-27 | | Release date: | 2023-10-18 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Molecular basis of Wnt biogenesis, secretion, and Wnt7-specific signaling.
Cell, 186, 2023
|
|
8TZP
 
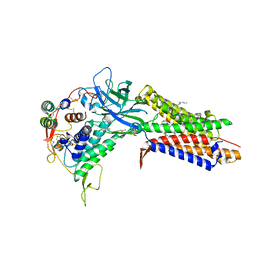 | | Structure of human Wnt7a bound to WLS and RECK | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, PALMITOLEIC ACID, Protein Wnt-7a, ... | | Authors: | Qi, X, Hu, Q, Li, X. | | Deposit date: | 2023-08-27 | | Release date: | 2023-10-18 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Molecular basis of Wnt biogenesis, secretion, and Wnt7-specific signaling.
Cell, 186, 2023
|
|
8TZR
 
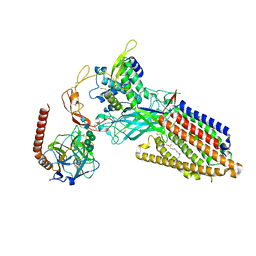 | | Structure of human Wnt3a bound to WLS and CALR | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Calreticulin, PALMITOLEIC ACID, ... | | Authors: | Qi, X, Hu, Q, Li, X. | | Deposit date: | 2023-08-27 | | Release date: | 2023-10-18 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of Wnt biogenesis, secretion, and Wnt7-specific signaling.
Cell, 186, 2023
|
|
8TZO
 
 | | Structure of human Wnt7a bound to WLS and CALR | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, Calreticulin, ... | | Authors: | Qi, X, Hu, Q, Li, X. | | Deposit date: | 2023-08-27 | | Release date: | 2023-10-18 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of Wnt biogenesis, secretion, and Wnt7-specific signaling.
Cell, 186, 2023
|
|
8U8J
 
 | |
8U8K
 
 | |
3M35
 
 | |
3MNV
 
 | | Crystal structure of the non-neutralizing HIV antibody 13H11 Fab fragment | | Descriptor: | 1,2-ETHANEDIOL, ANTI-HIV-1 ANTIBODY 13H11 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 13H11 LIGHT CHAIN, ... | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a non-neutralizing antibody to the HIV-1 gp41 membrane-proximal external region.
Nat.Struct.Mol.Biol., 17, 2010
|
|
